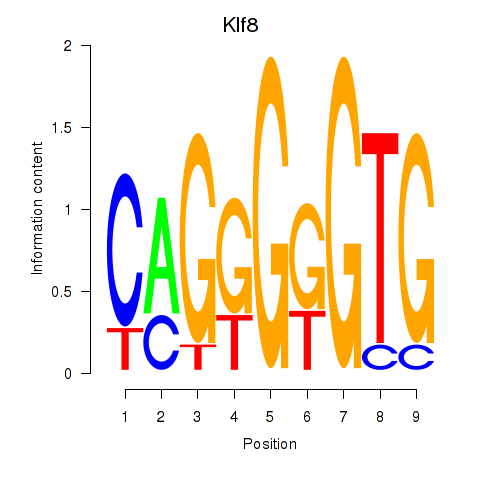
| 2.0 |
5.9 |
GO:0046022 |
regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 1.4 |
4.3 |
GO:1903795 |
regulation of inorganic anion transmembrane transport(GO:1903795) |
| 1.1 |
3.3 |
GO:0007521 |
muscle cell fate determination(GO:0007521) mammary placode formation(GO:0060596) |
| 1.1 |
3.3 |
GO:0060598 |
dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 1.0 |
5.9 |
GO:0072513 |
positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 1.0 |
1.9 |
GO:0016115 |
terpenoid catabolic process(GO:0016115) |
| 0.9 |
2.7 |
GO:0010725 |
regulation of primitive erythrocyte differentiation(GO:0010725) |
| 0.9 |
3.6 |
GO:0032439 |
endosome localization(GO:0032439) |
| 0.9 |
4.5 |
GO:0006369 |
termination of RNA polymerase II transcription(GO:0006369) |
| 0.8 |
2.5 |
GO:0035880 |
embryonic nail plate morphogenesis(GO:0035880) |
| 0.8 |
13.2 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.8 |
2.3 |
GO:0036166 |
phenotypic switching(GO:0036166) |
| 0.7 |
2.2 |
GO:1903225 |
negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.7 |
1.4 |
GO:1903224 |
regulation of endodermal cell fate specification(GO:0042663) regulation of endodermal cell differentiation(GO:1903224) |
| 0.7 |
2.0 |
GO:2000418 |
positive regulation of eosinophil migration(GO:2000418) |
| 0.7 |
2.0 |
GO:1904274 |
tricellular tight junction assembly(GO:1904274) |
| 0.6 |
4.4 |
GO:0000320 |
re-entry into mitotic cell cycle(GO:0000320) |
| 0.6 |
2.3 |
GO:0046836 |
glycolipid transport(GO:0046836) |
| 0.6 |
3.5 |
GO:0060242 |
contact inhibition(GO:0060242) |
| 0.5 |
2.7 |
GO:1903553 |
positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.5 |
1.6 |
GO:0036388 |
pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.5 |
2.1 |
GO:1904451 |
regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.5 |
1.0 |
GO:0061349 |
planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) |
| 0.5 |
5.3 |
GO:0048385 |
regulation of retinoic acid receptor signaling pathway(GO:0048385) |
| 0.5 |
0.5 |
GO:0070094 |
positive regulation of glucagon secretion(GO:0070094) |
| 0.5 |
2.4 |
GO:0018002 |
N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.4 |
1.8 |
GO:0042276 |
error-prone translesion synthesis(GO:0042276) |
| 0.4 |
1.3 |
GO:0002408 |
myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.4 |
1.7 |
GO:2000054 |
regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.4 |
4.5 |
GO:0042487 |
regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.4 |
2.0 |
GO:0046598 |
positive regulation of viral entry into host cell(GO:0046598) |
| 0.4 |
2.0 |
GO:0051045 |
negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.4 |
1.6 |
GO:0070345 |
negative regulation of fat cell proliferation(GO:0070345) |
| 0.4 |
2.7 |
GO:2001185 |
regulation of CD8-positive, alpha-beta T cell activation(GO:2001185) |
| 0.4 |
3.0 |
GO:0038028 |
insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.4 |
1.9 |
GO:0046864 |
retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) alveolar primary septum development(GO:0061143) |
| 0.4 |
1.1 |
GO:0030167 |
proteoglycan catabolic process(GO:0030167) |
| 0.4 |
1.8 |
GO:0071394 |
cellular response to testosterone stimulus(GO:0071394) |
| 0.3 |
2.3 |
GO:0072539 |
T-helper 17 cell differentiation(GO:0072539) |
| 0.3 |
1.6 |
GO:0010624 |
regulation of Schwann cell proliferation(GO:0010624) |
| 0.3 |
1.9 |
GO:0016576 |
histone dephosphorylation(GO:0016576) |
| 0.3 |
1.3 |
GO:0048143 |
astrocyte activation(GO:0048143) |
| 0.3 |
0.9 |
GO:0070844 |
misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.3 |
1.2 |
GO:0006842 |
tricarboxylic acid transport(GO:0006842) succinate transport(GO:0015744) citrate transport(GO:0015746) |
| 0.3 |
1.4 |
GO:2000507 |
positive regulation of energy homeostasis(GO:2000507) |
| 0.3 |
1.1 |
GO:0007056 |
spindle assembly involved in female meiosis(GO:0007056) |
| 0.3 |
0.8 |
GO:0007494 |
midgut development(GO:0007494) negative regulation of neuron maturation(GO:0014043) vein smooth muscle contraction(GO:0014826) |
| 0.2 |
1.0 |
GO:0060741 |
prostate gland stromal morphogenesis(GO:0060741) |
| 0.2 |
0.7 |
GO:2000402 |
negative regulation of lymphocyte migration(GO:2000402) |
| 0.2 |
1.6 |
GO:0016480 |
negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.2 |
3.7 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.2 |
0.2 |
GO:1904426 |
positive regulation of GTP binding(GO:1904426) |
| 0.2 |
0.7 |
GO:0042420 |
dopamine catabolic process(GO:0042420) |
| 0.2 |
2.2 |
GO:0042473 |
outer ear morphogenesis(GO:0042473) |
| 0.2 |
1.1 |
GO:0089700 |
positive regulation of T cell receptor signaling pathway(GO:0050862) protein kinase D signaling(GO:0089700) |
| 0.2 |
1.9 |
GO:0032286 |
central nervous system myelin maintenance(GO:0032286) |
| 0.2 |
0.6 |
GO:0010986 |
positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.2 |
0.8 |
GO:0060266 |
positive regulation of respiratory burst involved in inflammatory response(GO:0060265) negative regulation of respiratory burst involved in inflammatory response(GO:0060266) negative regulation of respiratory burst(GO:0060268) |
| 0.2 |
1.3 |
GO:0001842 |
neural fold formation(GO:0001842) |
| 0.2 |
0.7 |
GO:2001271 |
negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.2 |
0.9 |
GO:0090666 |
scaRNA localization to Cajal body(GO:0090666) |
| 0.2 |
0.6 |
GO:0014005 |
microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.2 |
0.9 |
GO:1900748 |
positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.2 |
1.6 |
GO:0051639 |
actin filament network formation(GO:0051639) |
| 0.2 |
0.7 |
GO:0035553 |
oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.2 |
0.9 |
GO:0006447 |
regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 0.2 |
0.8 |
GO:0030916 |
otic vesicle formation(GO:0030916) |
| 0.2 |
0.5 |
GO:0045041 |
protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.2 |
1.1 |
GO:0021913 |
regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.2 |
1.1 |
GO:0000103 |
sulfate assimilation(GO:0000103) |
| 0.1 |
0.4 |
GO:0071492 |
cellular response to UV-A(GO:0071492) |
| 0.1 |
0.3 |
GO:0043369 |
CD4-positive or CD8-positive, alpha-beta T cell lineage commitment(GO:0043369) |
| 0.1 |
1.3 |
GO:0060662 |
tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.1 |
4.5 |
GO:0009299 |
mRNA transcription(GO:0009299) |
| 0.1 |
0.8 |
GO:2000491 |
positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 |
1.1 |
GO:1903204 |
negative regulation of oxidative stress-induced neuron death(GO:1903204) |
| 0.1 |
1.6 |
GO:0070633 |
transepithelial transport(GO:0070633) |
| 0.1 |
1.8 |
GO:0038166 |
angiotensin-activated signaling pathway(GO:0038166) |
| 0.1 |
0.6 |
GO:2000623 |
regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 |
1.8 |
GO:0051451 |
myoblast migration(GO:0051451) |
| 0.1 |
4.3 |
GO:1900087 |
positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 |
1.0 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.1 |
1.4 |
GO:0046697 |
decidualization(GO:0046697) |
| 0.1 |
1.7 |
GO:0043011 |
myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 |
1.2 |
GO:0071670 |
smooth muscle cell chemotaxis(GO:0071670) |
| 0.1 |
0.4 |
GO:0032511 |
late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 |
0.7 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) |
| 0.1 |
0.4 |
GO:0040032 |
post-embryonic body morphogenesis(GO:0040032) |
| 0.1 |
1.2 |
GO:0099625 |
regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) ventricular cardiac muscle cell membrane repolarization(GO:0099625) |
| 0.1 |
0.4 |
GO:0035461 |
vitamin transmembrane transport(GO:0035461) |
| 0.1 |
1.4 |
GO:2000353 |
positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 |
1.2 |
GO:0042182 |
ketone catabolic process(GO:0042182) |
| 0.1 |
0.3 |
GO:2000271 |
positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 |
0.7 |
GO:0042711 |
maternal behavior(GO:0042711) |
| 0.1 |
1.6 |
GO:0015812 |
gamma-aminobutyric acid transport(GO:0015812) |
| 0.1 |
0.5 |
GO:0042135 |
neurotransmitter catabolic process(GO:0042135) |
| 0.1 |
1.2 |
GO:0048096 |
chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 |
0.8 |
GO:0060732 |
positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.1 |
1.2 |
GO:0038092 |
nodal signaling pathway(GO:0038092) |
| 0.1 |
0.3 |
GO:0031509 |
telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.1 |
1.1 |
GO:0035721 |
intraciliary retrograde transport(GO:0035721) |
| 0.1 |
0.7 |
GO:0051561 |
positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.1 |
1.0 |
GO:0002467 |
germinal center formation(GO:0002467) |
| 0.1 |
0.7 |
GO:0045647 |
negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 |
1.2 |
GO:0010640 |
regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) |
| 0.1 |
0.5 |
GO:0010571 |
positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 |
2.1 |
GO:0006270 |
DNA replication initiation(GO:0006270) |
| 0.1 |
1.4 |
GO:0090005 |
negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.1 |
1.5 |
GO:0045662 |
negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 |
2.9 |
GO:1902476 |
chloride transmembrane transport(GO:1902476) |
| 0.1 |
1.9 |
GO:0030261 |
chromosome condensation(GO:0030261) |
| 0.1 |
2.0 |
GO:0018345 |
protein palmitoylation(GO:0018345) |
| 0.1 |
0.3 |
GO:0014886 |
transition between slow and fast fiber(GO:0014886) |
| 0.1 |
1.5 |
GO:0060216 |
definitive hemopoiesis(GO:0060216) |
| 0.1 |
0.6 |
GO:0051386 |
regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.1 |
1.8 |
GO:0030513 |
positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 |
1.5 |
GO:0006346 |
methylation-dependent chromatin silencing(GO:0006346) |
| 0.1 |
0.8 |
GO:0006516 |
glycoprotein catabolic process(GO:0006516) |
| 0.1 |
0.5 |
GO:2001214 |
negative regulation of vascular permeability(GO:0043116) positive regulation of vasculogenesis(GO:2001214) |
| 0.1 |
0.3 |
GO:0097368 |
establishment of Sertoli cell barrier(GO:0097368) |
| 0.1 |
1.0 |
GO:0060033 |
anatomical structure regression(GO:0060033) |
| 0.1 |
0.2 |
GO:0015910 |
peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 |
0.7 |
GO:0046498 |
S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 |
0.2 |
GO:0009597 |
detection of virus(GO:0009597) |
| 0.0 |
0.5 |
GO:0061179 |
negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 |
0.6 |
GO:1903963 |
arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 |
0.2 |
GO:0060903 |
positive regulation of meiosis I(GO:0060903) |
| 0.0 |
1.4 |
GO:0006783 |
heme biosynthetic process(GO:0006783) |
| 0.0 |
1.0 |
GO:0030212 |
hyaluronan metabolic process(GO:0030212) |
| 0.0 |
1.7 |
GO:0045599 |
negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 |
0.6 |
GO:0032287 |
peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 |
0.7 |
GO:0010666 |
positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.0 |
0.3 |
GO:0007288 |
sperm axoneme assembly(GO:0007288) positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 |
0.8 |
GO:0071470 |
cellular response to osmotic stress(GO:0071470) |
| 0.0 |
0.2 |
GO:0060019 |
radial glial cell differentiation(GO:0060019) |
| 0.0 |
0.3 |
GO:0043923 |
positive regulation by host of viral transcription(GO:0043923) |
| 0.0 |
1.2 |
GO:0048791 |
calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 |
2.8 |
GO:0006096 |
glycolytic process(GO:0006096) ATP generation from ADP(GO:0006757) |
| 0.0 |
0.2 |
GO:0007197 |
adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 |
0.1 |
GO:0060750 |
epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) branch elongation involved in mammary gland duct branching(GO:0060751) |
| 0.0 |
1.2 |
GO:0000184 |
nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 |
1.9 |
GO:0007019 |
microtubule depolymerization(GO:0007019) |
| 0.0 |
0.4 |
GO:0016926 |
protein desumoylation(GO:0016926) |
| 0.0 |
0.2 |
GO:2000210 |
positive regulation of anoikis(GO:2000210) |
| 0.0 |
0.2 |
GO:0006362 |
transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 |
2.1 |
GO:0050709 |
negative regulation of protein secretion(GO:0050709) |
| 0.0 |
0.5 |
GO:0016486 |
peptide hormone processing(GO:0016486) |
| 0.0 |
0.0 |
GO:0002901 |
mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 |
0.3 |
GO:1903427 |
negative regulation of reactive oxygen species biosynthetic process(GO:1903427) |
| 0.0 |
0.8 |
GO:0042073 |
intraciliary transport(GO:0042073) |
| 0.0 |
1.3 |
GO:0031338 |
regulation of vesicle fusion(GO:0031338) |
| 0.0 |
0.7 |
GO:2000649 |
regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 |
0.4 |
GO:0016180 |
snRNA processing(GO:0016180) |
| 0.0 |
0.7 |
GO:0010501 |
RNA secondary structure unwinding(GO:0010501) |
| 0.0 |
1.0 |
GO:0006446 |
regulation of translational initiation(GO:0006446) |
| 0.0 |
0.8 |
GO:0030512 |
negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 |
0.3 |
GO:0030318 |
melanocyte differentiation(GO:0030318) |
| 0.0 |
0.7 |
GO:2001238 |
positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 |
0.3 |
GO:0043984 |
histone H4-K16 acetylation(GO:0043984) |
| 0.0 |
1.4 |
GO:0017148 |
negative regulation of translation(GO:0017148) |
| 0.0 |
0.6 |
GO:0006506 |
GPI anchor biosynthetic process(GO:0006506) |
| 0.0 |
0.7 |
GO:0032543 |
mitochondrial translation(GO:0032543) |
| 0.0 |
0.5 |
GO:0002181 |
cytoplasmic translation(GO:0002181) |
| 0.0 |
0.7 |
GO:0006073 |
glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 |
1.2 |
GO:0006633 |
fatty acid biosynthetic process(GO:0006633) |