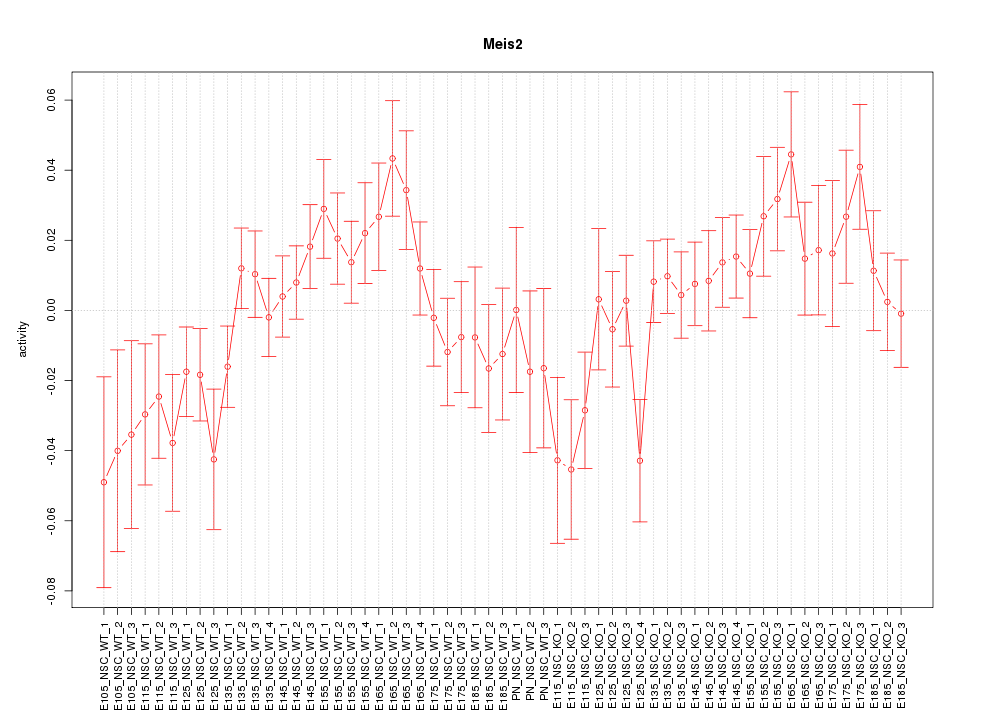
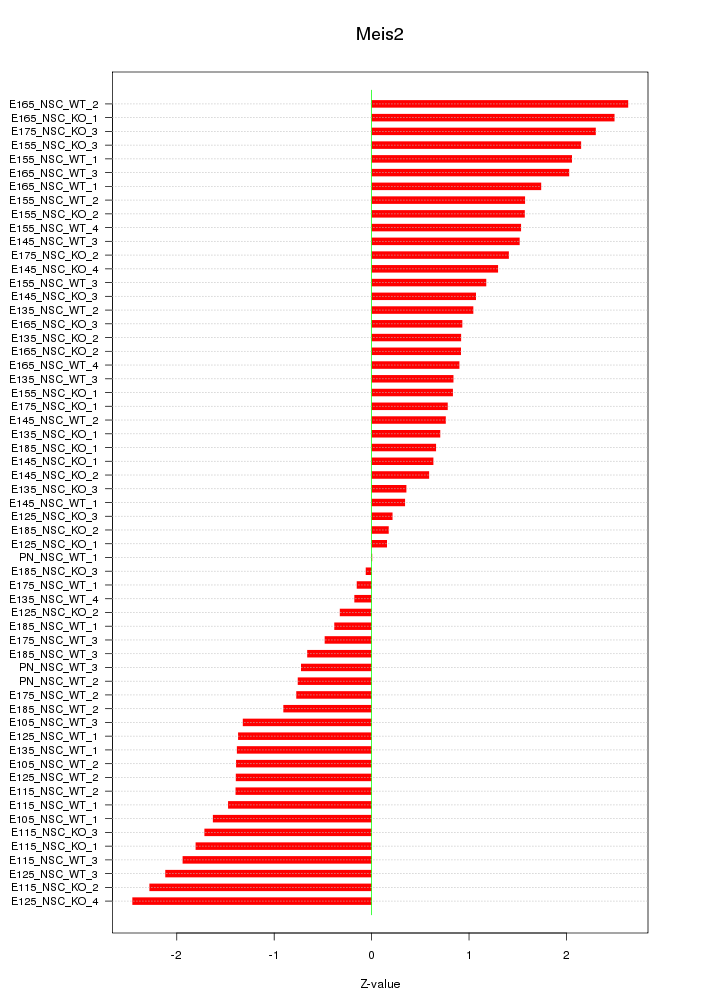
Motif ID: Meis2
Z-value: 1.332
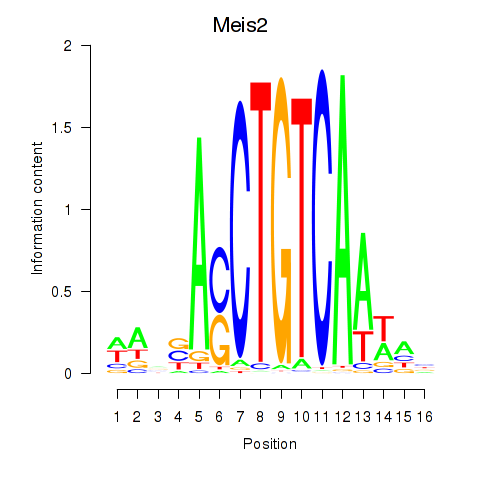
Transcription factors associated with Meis2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Meis2 | ENSMUSG00000027210.14 | Meis2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Meis2 | mm10_v2_chr2_-_116065798_116065853 | 0.68 | 3.3e-09 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.1 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 2.5 | 10.1 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 2.3 | 11.6 | GO:0019732 | antifungal humoral response(GO:0019732) |
| 1.9 | 7.6 | GO:0021564 | glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 1.6 | 4.8 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 1.5 | 8.9 | GO:0035743 | CD4-positive, alpha-beta T cell cytokine production(GO:0035743) T-helper 2 cell cytokine production(GO:0035745) |
| 1.4 | 14.2 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 1.4 | 7.1 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 1.3 | 4.0 | GO:0031038 | myosin II filament organization(GO:0031038) regulation of myosin II filament organization(GO:0043519) |
| 1.3 | 7.7 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.9 | 3.6 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.9 | 6.9 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.9 | 2.6 | GO:0071550 | regulation of muscle atrophy(GO:0014735) death-inducing signaling complex assembly(GO:0071550) negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.8 | 2.5 | GO:0042196 | dichloromethane metabolic process(GO:0018900) chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 0.8 | 8.4 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.7 | 7.5 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.7 | 3.6 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.7 | 3.4 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.6 | 10.3 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.6 | 3.6 | GO:1903887 | motile primary cilium assembly(GO:1903887) |
| 0.6 | 5.2 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) regulation of ribonucleoprotein complex localization(GO:2000197) |
| 0.6 | 5.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.6 | 1.7 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.5 | 1.6 | GO:0070428 | granuloma formation(GO:0002432) negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) toll-like receptor 5 signaling pathway(GO:0034146) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.5 | 4.9 | GO:0098907 | regulation of SA node cell action potential(GO:0098907) |
| 0.5 | 2.3 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.4 | 3.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.4 | 1.8 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.4 | 4.8 | GO:0071225 | cellular response to muramyl dipeptide(GO:0071225) |
| 0.4 | 3.9 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.4 | 5.4 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.4 | 7.3 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.4 | 3.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.4 | 1.6 | GO:0001907 | killing by symbiont of host cells(GO:0001907) induction of programmed cell death(GO:0012502) disruption by symbiont of host cell(GO:0044004) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.4 | 8.5 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.4 | 4.1 | GO:1901621 | regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901620) negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.4 | 8.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.4 | 1.8 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.3 | 1.7 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.3 | 12.5 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.3 | 1.6 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.3 | 1.3 | GO:0060046 | regulation of acrosome reaction(GO:0060046) |
| 0.3 | 2.2 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.3 | 7.7 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.3 | 1.5 | GO:0032423 | regulation of mismatch repair(GO:0032423) |
| 0.3 | 5.4 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.3 | 2.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.3 | 1.8 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.3 | 1.4 | GO:0097343 | positive regulation of T cell receptor signaling pathway(GO:0050862) ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.3 | 4.5 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.3 | 1.9 | GO:0044144 | regulation of growth of symbiont in host(GO:0044126) modulation of growth of symbiont involved in interaction with host(GO:0044144) |
| 0.3 | 1.6 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.3 | 3.7 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.3 | 3.4 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.3 | 1.3 | GO:0010730 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.2 | 2.7 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.2 | 2.5 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.2 | 0.7 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.2 | 1.2 | GO:0042538 | hyperosmotic salinity response(GO:0042538) |
| 0.2 | 0.7 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.2 | 1.1 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.2 | 0.9 | GO:1990564 | protein ufmylation(GO:0071569) protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.2 | 2.2 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.2 | 5.5 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.2 | 4.3 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.2 | 0.6 | GO:2000598 | regulation of cyclin catabolic process(GO:2000598) negative regulation of cyclin catabolic process(GO:2000599) |
| 0.2 | 0.6 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.2 | 1.1 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.2 | 1.5 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.2 | 4.5 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.2 | 0.7 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.2 | 0.9 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.2 | 1.9 | GO:0048070 | regulation of developmental pigmentation(GO:0048070) |
| 0.2 | 3.4 | GO:0032438 | melanosome organization(GO:0032438) anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.2 | 1.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 2.9 | GO:0001504 | neurotransmitter uptake(GO:0001504) |
| 0.2 | 0.5 | GO:0042701 | progesterone secretion(GO:0042701) |
| 0.2 | 2.6 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.2 | 4.1 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.2 | 4.5 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.1 | 0.6 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.1 | 0.7 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.1 | 2.2 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 1.1 | GO:0097460 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) ferrous iron import into cell(GO:0097460) |
| 0.1 | 0.5 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.1 | 0.6 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.1 | 1.1 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 0.6 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.1 | 3.3 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.1 | 9.6 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 3.6 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 1.9 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.1 | 1.6 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 0.7 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.1 | 6.3 | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.1 | 2.9 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 0.5 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.1 | 2.3 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.1 | 2.3 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.1 | 0.6 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.1 | 1.2 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 2.7 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.1 | 0.6 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.1 | 0.9 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 3.6 | GO:0048839 | inner ear development(GO:0048839) |
| 0.1 | 0.5 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 | 0.5 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.1 | 0.1 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.1 | 0.8 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 3.0 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.1 | 4.2 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.1 | 0.8 | GO:0030262 | cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.1 | 2.2 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 7.7 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 3.9 | GO:0060349 | bone morphogenesis(GO:0060349) |
| 0.0 | 1.9 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.3 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.9 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.5 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 4.6 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 2.4 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.4 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 2.3 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 4.2 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 1.8 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.4 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 2.4 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.0 | 1.1 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 4.6 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 0.1 | GO:0051683 | establishment of Golgi localization(GO:0051683) |
| 0.0 | 0.9 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 0.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.4 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 2.1 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 0.1 | GO:0006533 | aspartate catabolic process(GO:0006533) D-amino acid catabolic process(GO:0019478) |
| 0.0 | 0.9 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 0.9 | GO:0043473 | pigmentation(GO:0043473) |
| 0.0 | 0.4 | GO:0090102 | cochlea development(GO:0090102) |
| 0.0 | 0.2 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.4 | GO:0007599 | blood coagulation(GO:0007596) hemostasis(GO:0007599) |
| 0.0 | 0.1 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.2 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 | 0.8 | GO:0006821 | chloride transport(GO:0006821) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 9.4 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 1.6 | 8.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 1.3 | 14.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 1.2 | 4.8 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 1.2 | 3.6 | GO:0016939 | kinesin II complex(GO:0016939) |
| 1.1 | 10.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 1.0 | 3.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.9 | 10.3 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.7 | 2.8 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.7 | 9.2 | GO:0043203 | axon hillock(GO:0043203) |
| 0.6 | 4.5 | GO:0005883 | neurofilament(GO:0005883) |
| 0.6 | 7.0 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.6 | 1.7 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.6 | 4.5 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.5 | 5.7 | GO:0061700 | Seh1-associated complex(GO:0035859) GATOR2 complex(GO:0061700) |
| 0.5 | 2.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.4 | 5.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.4 | 6.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.4 | 3.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.4 | 4.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.3 | 4.8 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.3 | 3.6 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.3 | 6.9 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.3 | 1.9 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.2 | 2.5 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.2 | 2.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.2 | 2.1 | GO:0042599 | lamellar body(GO:0042599) |
| 0.2 | 0.9 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.2 | 1.2 | GO:0031314 | mitochondrial inner membrane presequence translocase complex(GO:0005744) extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.2 | 3.4 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.2 | 4.4 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 0.7 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.2 | 1.8 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.2 | 15.4 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 1.5 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.1 | 9.7 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 15.2 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 2.1 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.1 | 2.8 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 2.3 | GO:0042641 | actomyosin(GO:0042641) |
| 0.1 | 0.5 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 1.9 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 12.9 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 1.2 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 1.2 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.1 | 7.3 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 0.6 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.6 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 1.1 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 0.6 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.1 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.4 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 1.2 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 3.6 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 3.0 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.4 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 2.0 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 3.9 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 3.1 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 3.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 3.5 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.5 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 9.6 | GO:0097060 | synaptic membrane(GO:0097060) |
| 0.0 | 3.4 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.9 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.4 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 7.0 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.9 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 3.6 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 0.6 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.4 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.6 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.3 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.5 | GO:0045171 | intercellular bridge(GO:0045171) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.7 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 1.6 | 8.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 1.4 | 5.4 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) |
| 1.2 | 7.5 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 1.2 | 3.6 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 1.2 | 3.6 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 1.1 | 10.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 1.0 | 14.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 1.0 | 3.9 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 1.0 | 4.8 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.9 | 4.6 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.9 | 18.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.8 | 2.5 | GO:0019120 | hydrolase activity, acting on acid halide bonds(GO:0016824) hydrolase activity, acting on acid halide bonds, in C-halide compounds(GO:0019120) alkylhalidase activity(GO:0047651) |
| 0.8 | 7.0 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.7 | 2.2 | GO:0015173 | hydrogen:amino acid symporter activity(GO:0005280) aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.7 | 4.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.7 | 3.4 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.7 | 2.7 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.6 | 2.6 | GO:0035877 | death effector domain binding(GO:0035877) caspase binding(GO:0089720) |
| 0.6 | 3.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.6 | 10.1 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.5 | 3.7 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.5 | 1.5 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.5 | 2.7 | GO:0070728 | leucine binding(GO:0070728) |
| 0.4 | 5.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.4 | 7.1 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.4 | 7.3 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.4 | 1.8 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.3 | 6.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.3 | 4.8 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.3 | 1.3 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.3 | 4.0 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.3 | 7.0 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.3 | 3.0 | GO:0052872 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) phenanthrene 9,10-monooxygenase activity(GO:0018636) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) tocotrienol omega-hydroxylase activity(GO:0052872) thalianol hydroxylase activity(GO:0080014) |
| 0.3 | 3.8 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.3 | 1.4 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.3 | 1.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.3 | 2.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.3 | 3.9 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.2 | 0.7 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.2 | 5.3 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.2 | 5.0 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.2 | 2.5 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.2 | 0.9 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.2 | 6.2 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.2 | 4.8 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.2 | 0.6 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.2 | 0.6 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.2 | 0.7 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.2 | 0.9 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.2 | 6.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.2 | 1.1 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.2 | 0.8 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.2 | 2.9 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.1 | 1.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.1 | 4.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 4.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 2.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 1.9 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 1.5 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 2.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 1.6 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.1 | 1.8 | GO:0001163 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.1 | 4.5 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 5.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 3.0 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 2.9 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.1 | 2.1 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 3.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 3.6 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 1.1 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 9.9 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.1 | 5.7 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 1.8 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 0.8 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 1.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 1.9 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 1.2 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 6.6 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 0.4 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 1.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 0.8 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 1.8 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.9 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 1.8 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 3.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 4.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 13.4 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 1.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.6 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.6 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.8 | GO:0052771 | coenzyme F390-A hydrolase activity(GO:0052770) coenzyme F390-G hydrolase activity(GO:0052771) |
| 0.0 | 0.7 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 1.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.2 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 1.1 | GO:0000009 | alpha-1,6-mannosyltransferase activity(GO:0000009) |
| 0.0 | 0.1 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) aspartate oxidase activity(GO:0015922) |
| 0.0 | 0.1 | GO:0103116 | alpha-D-galactofuranose transporter activity(GO:0103116) |
| 0.0 | 0.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 1.2 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.0 | 2.6 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.1 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) |
| 0.0 | 0.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.4 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.5 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |