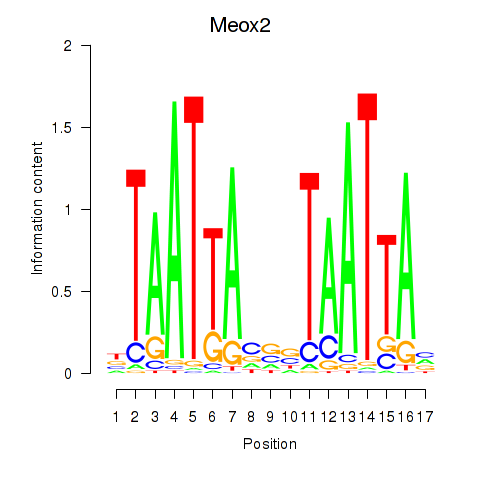
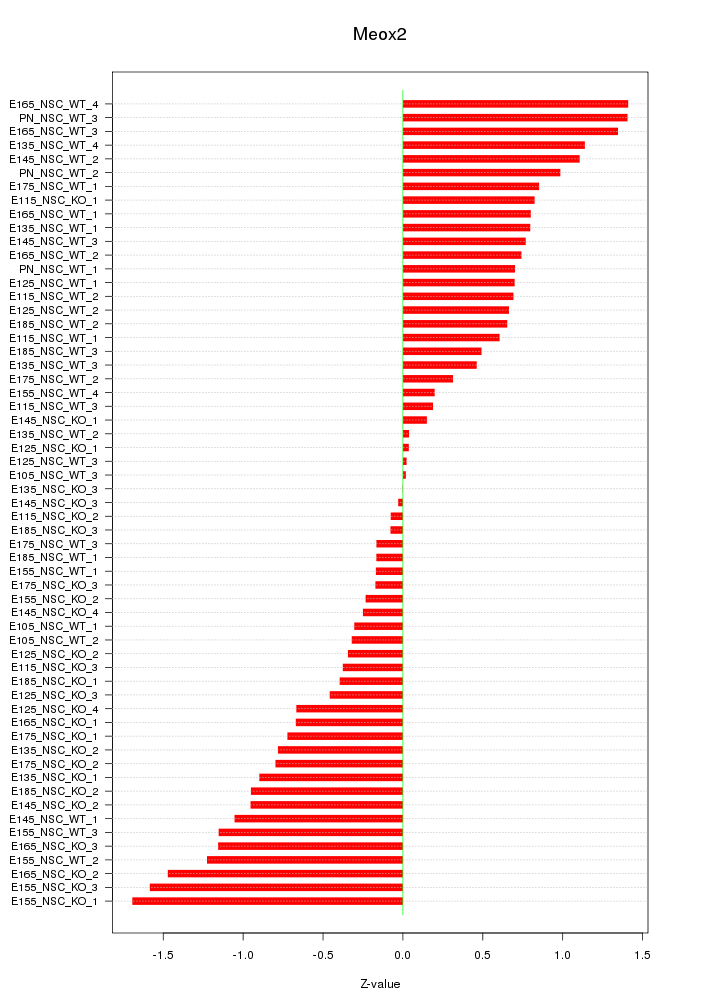
| 0.6 |
1.8 |
GO:0006566 |
threonine metabolic process(GO:0006566) |
| 0.5 |
1.6 |
GO:0032058 |
positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.5 |
1.5 |
GO:0008228 |
opsonization(GO:0008228) modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.4 |
1.3 |
GO:0060126 |
somatotropin secreting cell differentiation(GO:0060126) |
| 0.4 |
1.1 |
GO:1902528 |
regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.3 |
6.0 |
GO:0097154 |
GABAergic neuron differentiation(GO:0097154) |
| 0.3 |
1.3 |
GO:0032788 |
saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.3 |
1.7 |
GO:0046602 |
regulation of mitotic centrosome separation(GO:0046602) |
| 0.3 |
1.1 |
GO:0000707 |
meiotic DNA recombinase assembly(GO:0000707) resolution of recombination intermediates(GO:0071139) |
| 0.2 |
1.0 |
GO:2000346 |
negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.2 |
1.0 |
GO:0035609 |
C-terminal protein deglutamylation(GO:0035609) |
| 0.2 |
1.2 |
GO:0003431 |
growth plate cartilage chondrocyte development(GO:0003431) |
| 0.2 |
1.5 |
GO:0000022 |
mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.1 |
0.8 |
GO:0010528 |
regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.1 |
0.8 |
GO:0006287 |
base-excision repair, gap-filling(GO:0006287) |
| 0.1 |
1.0 |
GO:0061158 |
3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 |
13.0 |
GO:0045580 |
regulation of T cell differentiation(GO:0045580) |
| 0.1 |
0.6 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.1 |
1.7 |
GO:0021521 |
ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 |
0.9 |
GO:1901970 |
positive regulation of mitotic sister chromatid separation(GO:1901970) |
| 0.1 |
1.8 |
GO:0090005 |
negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.1 |
1.1 |
GO:1903077 |
negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.1 |
0.5 |
GO:0007128 |
meiotic prophase I(GO:0007128) prophase(GO:0051324) meiotic cell cycle phase(GO:0098762) meiosis I cell cycle phase(GO:0098764) |
| 0.1 |
0.9 |
GO:2001214 |
positive regulation of vasculogenesis(GO:2001214) |
| 0.1 |
0.3 |
GO:0003406 |
retinal pigment epithelium development(GO:0003406) |
| 0.1 |
1.6 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 |
2.7 |
GO:0010862 |
positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.1 |
0.4 |
GO:0033762 |
response to glucagon(GO:0033762) |
| 0.1 |
1.2 |
GO:0001574 |
ganglioside biosynthetic process(GO:0001574) |
| 0.1 |
1.9 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.1 |
0.6 |
GO:0003417 |
growth plate cartilage development(GO:0003417) |
| 0.1 |
0.2 |
GO:0016598 |
protein arginylation(GO:0016598) |
| 0.0 |
0.4 |
GO:0051775 |
response to redox state(GO:0051775) |
| 0.0 |
0.8 |
GO:0034453 |
microtubule anchoring(GO:0034453) |
| 0.0 |
1.3 |
GO:0072698 |
protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 |
0.2 |
GO:2000325 |
regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.0 |
1.0 |
GO:0007271 |
synaptic transmission, cholinergic(GO:0007271) |
| 0.0 |
0.4 |
GO:0021796 |
cerebral cortex regionalization(GO:0021796) |
| 0.0 |
0.8 |
GO:0001946 |
lymphangiogenesis(GO:0001946) lymph vessel morphogenesis(GO:0036303) |
| 0.0 |
0.8 |
GO:0000413 |
protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 |
0.3 |
GO:0071763 |
nuclear membrane organization(GO:0071763) |
| 0.0 |
1.3 |
GO:0009225 |
nucleotide-sugar metabolic process(GO:0009225) |
| 0.0 |
0.3 |
GO:0031937 |
positive regulation of chromatin silencing(GO:0031937) |
| 0.0 |
0.9 |
GO:0006270 |
DNA replication initiation(GO:0006270) |
| 0.0 |
0.6 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 |
0.3 |
GO:0006953 |
acute-phase response(GO:0006953) |
| 0.0 |
0.1 |
GO:0009162 |
deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.0 |
0.5 |
GO:0032467 |
positive regulation of cytokinesis(GO:0032467) |
| 0.0 |
1.6 |
GO:0008543 |
fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 |
1.2 |
GO:0010923 |
negative regulation of phosphatase activity(GO:0010923) |
| 0.0 |
0.0 |
GO:0006041 |
glucosamine metabolic process(GO:0006041) |
| 0.0 |
0.0 |
GO:0006419 |
alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 |
0.4 |
GO:0007608 |
sensory perception of smell(GO:0007608) |
| 0.0 |
0.2 |
GO:0032967 |
positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 |
0.3 |
GO:0008340 |
determination of adult lifespan(GO:0008340) |
| 0.0 |
0.1 |
GO:0030539 |
male genitalia development(GO:0030539) |