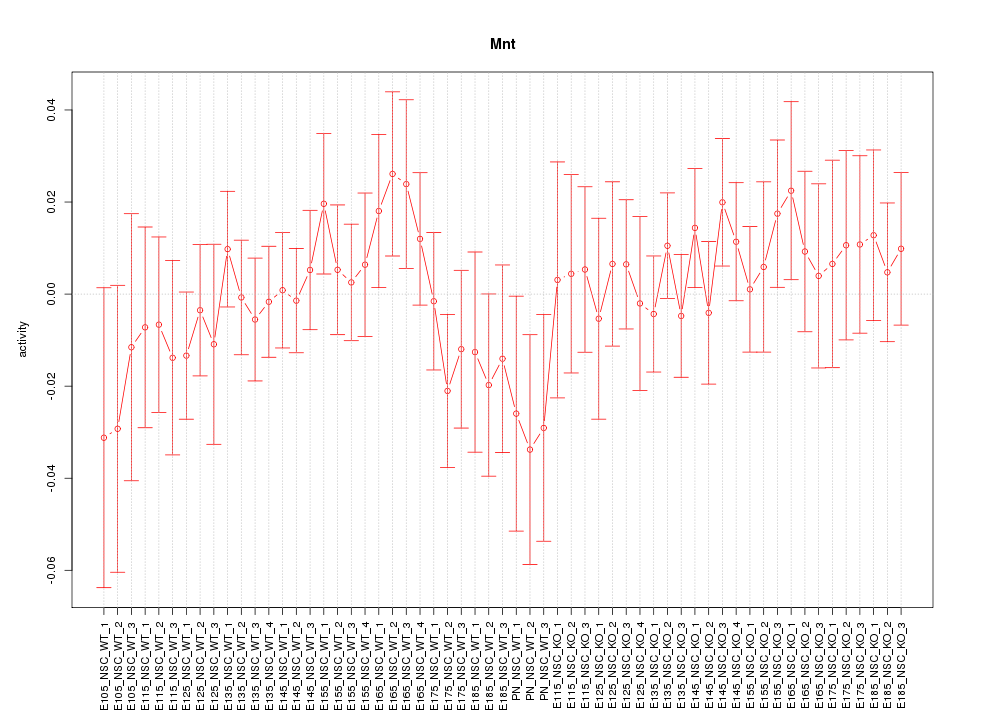
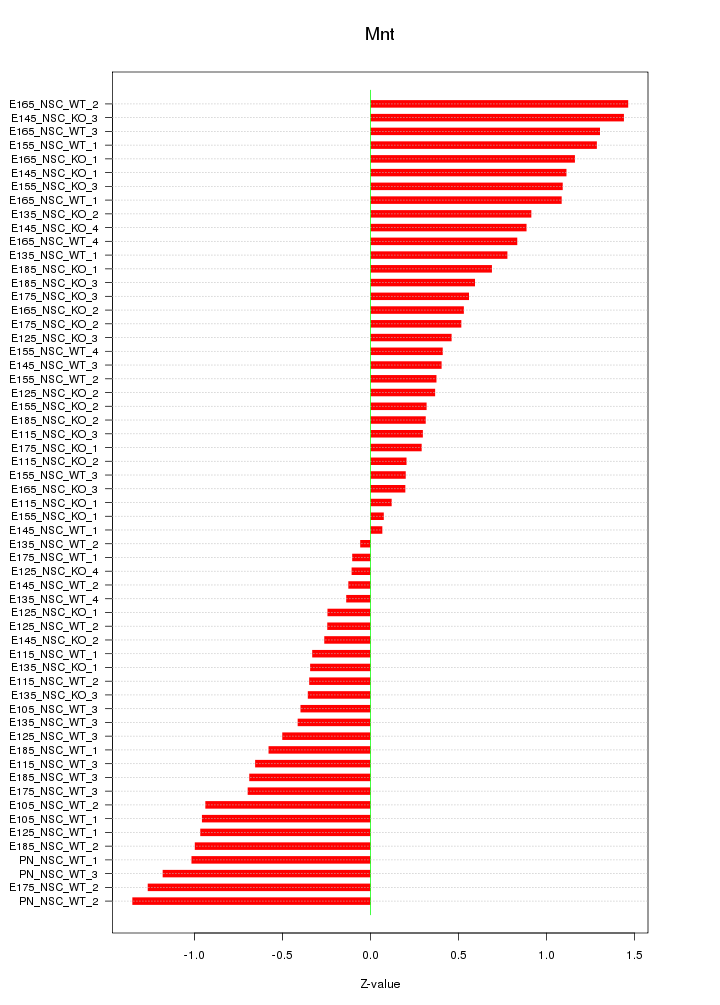
Motif ID: Mnt
Z-value: 0.726
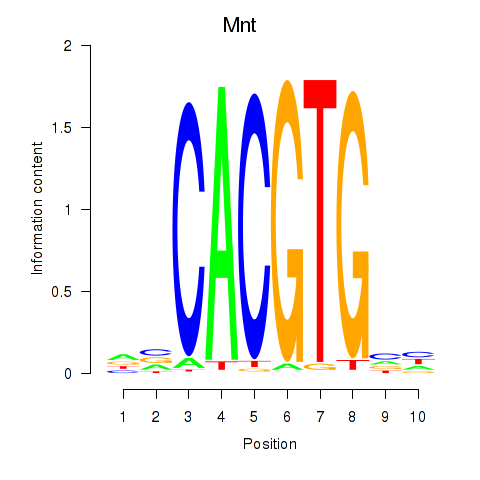
Transcription factors associated with Mnt:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Mnt | ENSMUSG00000000282.6 | Mnt |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mnt | mm10_v2_chr11_+_74830920_74831005 | 0.72 | 1.2e-10 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:0043379 | memory T cell differentiation(GO:0043379) |
| 1.5 | 1.5 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 1.4 | 4.2 | GO:1904049 | regulation of spontaneous neurotransmitter secretion(GO:1904048) negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 1.2 | 4.9 | GO:0021586 | pons maturation(GO:0021586) |
| 1.1 | 3.4 | GO:0001978 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) |
| 1.1 | 3.2 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 1.1 | 4.2 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.9 | 5.3 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.9 | 5.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.8 | 2.5 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.8 | 2.4 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.8 | 4.0 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.8 | 2.3 | GO:0070428 | granuloma formation(GO:0002432) negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) toll-like receptor 5 signaling pathway(GO:0034146) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.8 | 2.3 | GO:0060023 | soft palate development(GO:0060023) |
| 0.7 | 2.2 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.7 | 3.6 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.7 | 5.0 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.7 | 6.9 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.7 | 2.1 | GO:0046881 | positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.7 | 2.7 | GO:0061428 | embryonic heart tube left/right pattern formation(GO:0060971) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.7 | 2.6 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.6 | 2.6 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.6 | 2.6 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.6 | 2.4 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.6 | 2.3 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.6 | 2.9 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.5 | 5.9 | GO:1901250 | regulation of lung goblet cell differentiation(GO:1901249) negative regulation of lung goblet cell differentiation(GO:1901250) |
| 0.5 | 2.6 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.5 | 1.9 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.5 | 1.9 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.5 | 1.9 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.5 | 2.3 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.5 | 0.9 | GO:0003383 | apical constriction(GO:0003383) |
| 0.4 | 3.0 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.4 | 2.5 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.4 | 1.2 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.4 | 6.0 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.4 | 3.6 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.4 | 3.5 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.4 | 2.4 | GO:0071638 | positive regulation of T-helper 1 cell differentiation(GO:0045627) negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.4 | 4.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.4 | 11.4 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.4 | 4.2 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.4 | 0.7 | GO:0060025 | regulation of synaptic activity(GO:0060025) |
| 0.4 | 2.2 | GO:0021636 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) |
| 0.4 | 2.2 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.4 | 3.3 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.4 | 2.2 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.4 | 1.1 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.3 | 1.4 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.3 | 1.0 | GO:0014877 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.3 | 3.0 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.3 | 1.3 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.3 | 1.7 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.3 | 1.4 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.3 | 3.0 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.3 | 8.7 | GO:0015991 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.3 | 0.8 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.3 | 0.8 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.3 | 1.8 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.3 | 2.3 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.3 | 1.0 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.3 | 4.0 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.3 | 0.5 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.2 | 1.5 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.2 | 0.5 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.2 | 0.7 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.2 | 1.6 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.2 | 0.7 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.2 | 2.9 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.2 | 0.9 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.2 | 1.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.2 | 0.6 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.2 | 0.6 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.2 | 2.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.2 | 3.8 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) |
| 0.2 | 3.3 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.2 | 1.4 | GO:1903441 | protein localization to ciliary membrane(GO:1903441) |
| 0.2 | 6.4 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.2 | 0.2 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 0.2 | 4.6 | GO:0097576 | autophagosome maturation(GO:0097352) vacuole fusion(GO:0097576) |
| 0.2 | 2.0 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.2 | 0.6 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.2 | 1.4 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.2 | 0.6 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.2 | 1.0 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.2 | 1.9 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) positive regulation of ERAD pathway(GO:1904294) |
| 0.2 | 0.5 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.2 | 0.9 | GO:0032423 | regulation of mismatch repair(GO:0032423) |
| 0.2 | 0.7 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.2 | 0.5 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.2 | 1.7 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.2 | 0.5 | GO:0006592 | ornithine biosynthetic process(GO:0006592) |
| 0.2 | 0.8 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.2 | 0.5 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.2 | 1.8 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 | 1.3 | GO:0015074 | DNA integration(GO:0015074) |
| 0.2 | 3.7 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.2 | 2.7 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.2 | 2.3 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.2 | 6.2 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 0.6 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 | 0.3 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.1 | 0.6 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.1 | 0.9 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.1 | 3.1 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.1 | 0.8 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.1 | 1.7 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 1.2 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.1 | 0.5 | GO:1902369 | negative regulation of RNA catabolic process(GO:1902369) |
| 0.1 | 1.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 0.8 | GO:2001012 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.1 | 2.6 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 2.3 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 1.9 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.1 | 1.2 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.1 | 1.9 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.1 | 1.2 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 0.7 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.1 | 0.4 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.1 | 1.4 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.1 | 0.6 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 | 0.8 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.1 | 1.1 | GO:1904152 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) regulation of retrograde protein transport, ER to cytosol(GO:1904152) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 0.4 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.1 | 2.0 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.1 | 0.6 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.1 | 0.8 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.1 | 0.5 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.1 | 1.9 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 0.8 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 3.2 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.1 | 2.4 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 2.6 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 0.7 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.1 | 0.7 | GO:0060693 | regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.1 | 0.9 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 1.6 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 | 0.6 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 0.5 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 3.7 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 1.4 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 0.4 | GO:0090148 | membrane fission(GO:0090148) |
| 0.1 | 0.3 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.1 | 1.7 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.1 | 0.2 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) myotome development(GO:0061055) |
| 0.1 | 0.9 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 1.0 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 1.2 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.1 | 1.2 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.1 | 1.5 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.1 | 2.5 | GO:0006024 | glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.1 | 0.4 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.1 | 0.3 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.1 | 1.8 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.1 | 0.3 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900175) positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.1 | 1.3 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.1 | 2.4 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.1 | 0.7 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.1 | 2.0 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 3.1 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 0.8 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.1 | 3.1 | GO:0008542 | visual learning(GO:0008542) |
| 0.1 | 1.2 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.1 | 1.5 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.1 | 1.1 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 0.3 | GO:0099515 | actin filament-based transport(GO:0099515) |
| 0.1 | 1.4 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.1 | 1.0 | GO:2000279 | negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.1 | 0.5 | GO:0060979 | vasculogenesis involved in coronary vascular morphogenesis(GO:0060979) |
| 0.1 | 0.7 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 3.8 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 0.2 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 1.4 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 0.5 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.6 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.1 | 0.3 | GO:0015911 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) fatty acid transmembrane transport(GO:1902001) |
| 0.1 | 1.6 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 3.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 1.1 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.1 | 2.6 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 2.1 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 | 0.7 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.3 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.7 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.5 | GO:1901409 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.2 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.8 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.3 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.5 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.2 | GO:0021506 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.0 | 0.5 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.0 | 0.1 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.0 | 1.6 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.4 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) |
| 0.0 | 0.5 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.2 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.0 | 0.4 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.4 | GO:0032402 | melanosome transport(GO:0032402) |
| 0.0 | 2.4 | GO:0007569 | cell aging(GO:0007569) |
| 0.0 | 4.9 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 1.1 | GO:0034243 | regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.0 | 0.4 | GO:0006085 | acetyl-CoA biosynthetic process(GO:0006085) |
| 0.0 | 0.5 | GO:0009081 | branched-chain amino acid metabolic process(GO:0009081) branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.2 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.7 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.6 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.3 | GO:0019348 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) dolichol metabolic process(GO:0019348) |
| 0.0 | 0.1 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.1 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.0 | 0.2 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.0 | 0.4 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 3.4 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 0.3 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 1.2 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 3.1 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 0.3 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.3 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.4 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 0.9 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.2 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.5 | GO:0018208 | peptidyl-proline modification(GO:0018208) |
| 0.0 | 0.4 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 0.2 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.1 | GO:0051495 | positive regulation of cytoskeleton organization(GO:0051495) |
| 0.0 | 0.5 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.0 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) |
| 0.0 | 0.6 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.2 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.0 | 0.6 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.4 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.1 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.4 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.1 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 1.0 | GO:0006109 | regulation of carbohydrate metabolic process(GO:0006109) |
| 0.0 | 0.8 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.2 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.5 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.9 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.8 | 4.2 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.7 | 6.7 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.7 | 2.0 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.6 | 4.0 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.5 | 2.6 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.5 | 2.1 | GO:0043511 | inhibin complex(GO:0043511) |
| 0.5 | 6.9 | GO:0043203 | axon hillock(GO:0043203) |
| 0.5 | 3.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.5 | 1.4 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.4 | 2.2 | GO:0000798 | nuclear cohesin complex(GO:0000798) nuclear meiotic cohesin complex(GO:0034991) |
| 0.4 | 1.2 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.3 | 0.7 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.3 | 2.6 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.3 | 2.2 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.3 | 3.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.3 | 0.9 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.3 | 5.6 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.3 | 2.2 | GO:0097433 | dense body(GO:0097433) |
| 0.3 | 5.5 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.3 | 2.8 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.2 | 1.0 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.2 | 0.9 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.2 | 1.6 | GO:0070695 | FHF complex(GO:0070695) |
| 0.2 | 1.3 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.2 | 1.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.2 | 2.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.2 | 4.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.2 | 1.6 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.2 | 1.9 | GO:0034709 | methylosome(GO:0034709) |
| 0.2 | 1.2 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.2 | 3.4 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.2 | 1.4 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.2 | 1.9 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.2 | 1.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.2 | 1.3 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.2 | 0.8 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.2 | 3.3 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.2 | 2.6 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 0.7 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 1.3 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 0.4 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 2.4 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.1 | 0.9 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 0.5 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.1 | 0.8 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 1.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 4.6 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 1.9 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 1.0 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.1 | 3.8 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 1.2 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 1.1 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 0.7 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.3 | GO:1990879 | CST complex(GO:1990879) |
| 0.1 | 1.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.9 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 1.8 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.4 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.1 | 1.4 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 5.1 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 1.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.9 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 2.2 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.3 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.1 | 3.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 1.0 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 0.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.5 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.1 | 2.3 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 0.4 | GO:0044308 | axonal spine(GO:0044308) |
| 0.1 | 0.5 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 0.6 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 0.6 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 0.9 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 3.7 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 1.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 1.0 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 0.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 0.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 4.3 | GO:0030117 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.0 | 0.5 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.3 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.6 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.4 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 1.3 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 9.6 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 2.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 2.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.8 | GO:0098797 | plasma membrane protein complex(GO:0098797) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.7 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.5 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 6.7 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 5.9 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 3.2 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 1.6 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.4 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.6 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.3 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.9 | GO:0097060 | synaptic membrane(GO:0097060) |
| 0.0 | 2.3 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 2.5 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 9.6 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 2.2 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.7 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 1.5 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 1.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) CIA complex(GO:0097361) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.6 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.0 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.5 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.2 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.1 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 0.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.9 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.3 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.2 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) nuclear transcriptional repressor complex(GO:0090568) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0031752 | D3 dopamine receptor binding(GO:0031750) D5 dopamine receptor binding(GO:0031752) |
| 0.9 | 3.5 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.8 | 2.5 | GO:0035175 | histone kinase activity (H3-S10 specific)(GO:0035175) |
| 0.6 | 1.9 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.6 | 3.2 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.6 | 2.3 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.6 | 2.9 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.6 | 2.2 | GO:0035851 | histone deacetylase activity (H4-K16 specific)(GO:0034739) Krueppel-associated box domain binding(GO:0035851) |
| 0.5 | 4.2 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.5 | 6.0 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.5 | 0.5 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.5 | 1.5 | GO:0071253 | connexin binding(GO:0071253) |
| 0.5 | 2.0 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.5 | 1.9 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.5 | 8.6 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.5 | 3.8 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.5 | 1.4 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.5 | 1.8 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.4 | 5.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.4 | 3.0 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.4 | 1.2 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.4 | 1.2 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.4 | 1.2 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.4 | 1.2 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.4 | 1.2 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.4 | 10.7 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.4 | 1.9 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.4 | 7.9 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.4 | 3.4 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.4 | 2.6 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.4 | 4.0 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.4 | 1.8 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.3 | 1.4 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.3 | 2.7 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.3 | 1.7 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.3 | 3.4 | GO:0004889 | acetylcholine-activated cation-selective channel activity(GO:0004889) |
| 0.3 | 0.9 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.3 | 1.3 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.3 | 2.9 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.3 | 2.6 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.3 | 0.8 | GO:0015173 | hydrogen:amino acid symporter activity(GO:0005280) aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.3 | 1.3 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.2 | 3.7 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.2 | 7.4 | GO:0034930 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) heparan sulfate 2-O-sulfotransferase activity(GO:0004394) HNK-1 sulfotransferase activity(GO:0016232) heparan sulfate 6-O-sulfotransferase activity(GO:0017095) trans-9R,10R-dihydrodiolphenanthrene sulfotransferase activity(GO:0018721) 1-phenanthrol sulfotransferase activity(GO:0018722) 3-phenanthrol sulfotransferase activity(GO:0018723) 4-phenanthrol sulfotransferase activity(GO:0018724) trans-3,4-dihydrodiolphenanthrene sulfotransferase activity(GO:0018725) 9-phenanthrol sulfotransferase activity(GO:0018726) 2-phenanthrol sulfotransferase activity(GO:0018727) phenanthrol sulfotransferase activity(GO:0019111) 1-hydroxypyrene sulfotransferase activity(GO:0034930) proteoglycan sulfotransferase activity(GO:0050698) cholesterol sulfotransferase activity(GO:0051922) hydroxyjasmonate sulfotransferase activity(GO:0080131) |
| 0.2 | 1.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.2 | 2.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.2 | 2.6 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.2 | 4.9 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.2 | 1.0 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.2 | 1.9 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.2 | 3.0 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.2 | 1.0 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.2 | 1.2 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.2 | 3.1 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 1.0 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.2 | 7.4 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.2 | 2.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 0.7 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.2 | 0.7 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.2 | 9.3 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.2 | 1.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.2 | 0.5 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) |
| 0.2 | 5.0 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.2 | 0.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.2 | 2.7 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.2 | 0.5 | GO:0017084 | glutamate 5-kinase activity(GO:0004349) glutamate-5-semialdehyde dehydrogenase activity(GO:0004350) delta1-pyrroline-5-carboxylate synthetase activity(GO:0017084) amino acid kinase activity(GO:0019202) |
| 0.2 | 0.5 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.2 | 0.5 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.2 | 2.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.2 | 0.6 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.2 | 4.2 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 4.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 0.4 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.1 | 3.1 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.4 | GO:0004980 | melanocortin receptor activity(GO:0004977) melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.1 | 3.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 1.1 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.1 | 2.9 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.1 | 4.2 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 0.6 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.1 | 0.8 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 2.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.7 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 0.4 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 1.9 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 3.1 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 2.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 2.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 2.6 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 1.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 0.4 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.1 | 2.0 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 0.6 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 2.5 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 0.5 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.1 | 1.3 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 0.5 | GO:0000339 | RNA cap binding(GO:0000339) translation activator activity(GO:0008494) |
| 0.1 | 2.0 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 1.8 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.1 | 2.0 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.1 | 0.8 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 0.5 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.1 | 0.5 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 1.0 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.3 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.1 | 2.4 | GO:0018602 | sulfonate dioxygenase activity(GO:0000907) 2,4-dichlorophenoxyacetate alpha-ketoglutarate dioxygenase activity(GO:0018602) hypophosphite dioxygenase activity(GO:0034792) gibberellin 2-beta-dioxygenase activity(GO:0045543) C-19 gibberellin 2-beta-dioxygenase activity(GO:0052634) C-20 gibberellin 2-beta-dioxygenase activity(GO:0052635) |
| 0.1 | 0.4 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 1.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 2.0 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 1.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.6 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 0.3 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 2.9 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 0.4 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 0.4 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 0.7 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 0.3 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 1.1 | GO:0035380 | C-3 sterol dehydrogenase (C-4 sterol decarboxylase) activity(GO:0000252) mevaldate reductase activity(GO:0004495) gluconate dehydrogenase activity(GO:0008875) epoxide dehydrogenase activity(GO:0018451) 5-exo-hydroxycamphor dehydrogenase activity(GO:0018452) 2-hydroxytetrahydrofuran dehydrogenase activity(GO:0018453) acetoin dehydrogenase activity(GO:0019152) phenylcoumaran benzylic ether reductase activity(GO:0032442) D-xylose:NADP reductase activity(GO:0032866) L-arabinose:NADP reductase activity(GO:0032867) D-arabinitol dehydrogenase, D-ribulose forming (NADP+) activity(GO:0033709) (R)-(-)-1,2,3,4-tetrahydronaphthol dehydrogenase activity(GO:0034831) 3-hydroxymenthone dehydrogenase activity(GO:0034840) very long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0035380) dihydrotestosterone 17-beta-dehydrogenase activity(GO:0035410) (R)-2-hydroxyisocaproate dehydrogenase activity(GO:0043713) L-arabinose 1-dehydrogenase (NADP+) activity(GO:0044103) L-xylulose reductase (NAD+) activity(GO:0044105) 3-ketoglucose-reductase activity(GO:0048258) D-arabinitol dehydrogenase, D-xylulose forming (NADP+) activity(GO:0052677) |
| 0.1 | 0.2 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.1 | 0.3 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 0.6 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 1.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.3 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.1 | 3.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.4 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 1.6 | GO:0045502 | dynein binding(GO:0045502) |
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.3 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.0 | 4.2 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.2 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 1.3 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.8 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 1.8 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.7 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.4 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 9.6 | GO:0043774 | UDP-N-acetylmuramoylalanyl-D-glutamyl-2,6-diaminopimelate-D-alanyl-D-alanine ligase activity(GO:0008766) ribosomal S6-glutamic acid ligase activity(GO:0018169) coenzyme F420-0 gamma-glutamyl ligase activity(GO:0043773) coenzyme F420-2 alpha-glutamyl ligase activity(GO:0043774) protein-glycine ligase activity(GO:0070735) protein-glycine ligase activity, initiating(GO:0070736) protein-glycine ligase activity, elongating(GO:0070737) tubulin-glycine ligase activity(GO:0070738) |
| 0.0 | 1.9 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.0 | 3.0 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.3 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 1.1 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 2.2 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 5.5 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.3 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.6 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.2 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 1.5 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.2 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 1.2 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 7.5 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 6.3 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 0.8 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.6 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 1.4 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 1.6 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 2.1 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.0 | 1.7 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.4 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.1 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.3 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.3 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.4 | GO:0019707 | protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.0 | 0.0 | GO:0008521 | acetyl-CoA transporter activity(GO:0008521) |
| 0.0 | 0.2 | GO:0005521 | lamin binding(GO:0005521) |