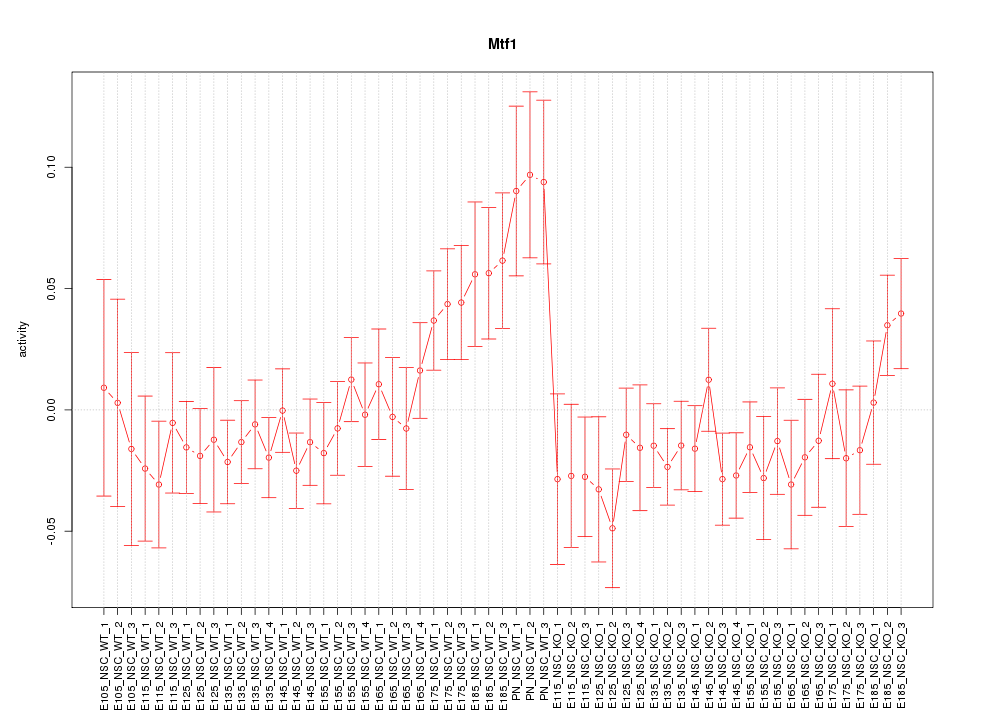
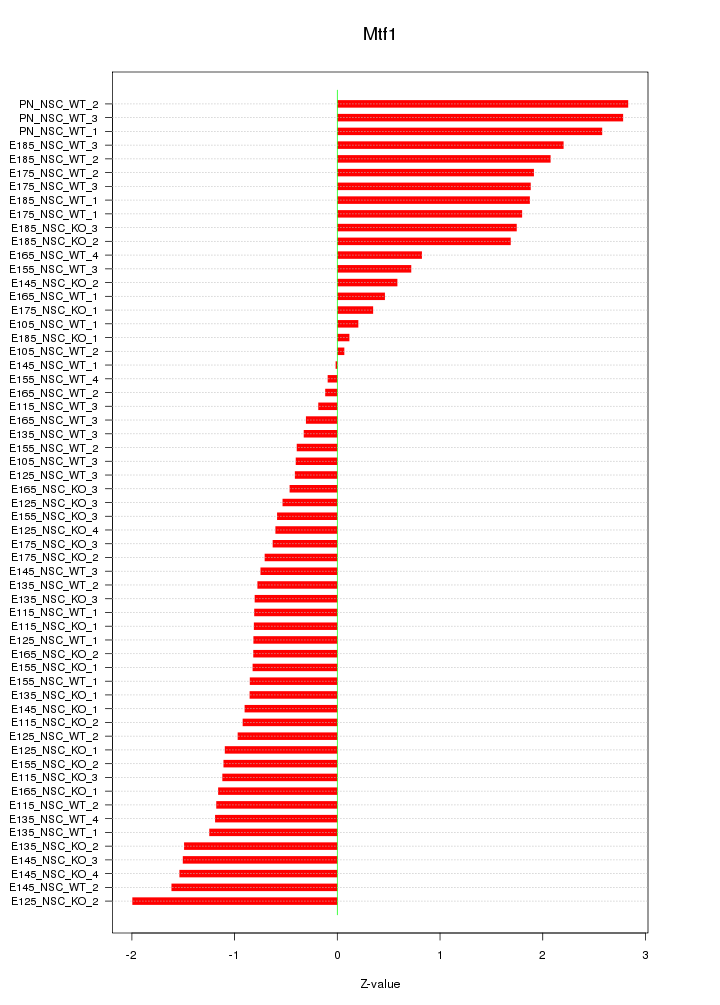
Motif ID: Mtf1
Z-value: 1.223
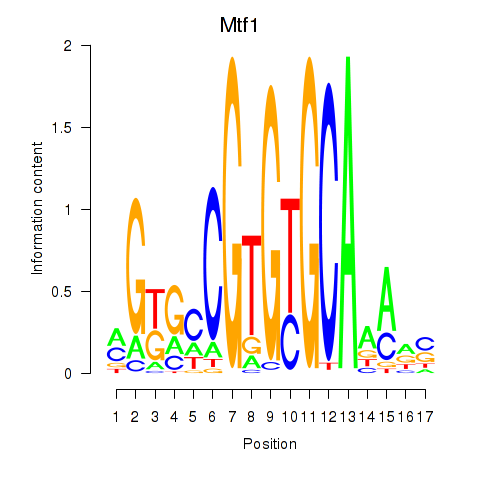
Transcription factors associated with Mtf1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Mtf1 | ENSMUSG00000028890.7 | Mtf1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mtf1 | mm10_v2_chr4_+_124802543_124802678 | 0.38 | 2.7e-03 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.0 | 70.3 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 8.6 | 8.6 | GO:0061687 | detoxification of inorganic compound(GO:0061687) |
| 1.6 | 29.0 | GO:0032060 | bleb assembly(GO:0032060) |
| 1.1 | 3.2 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.9 | 5.6 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.9 | 6.2 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.9 | 2.6 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.8 | 5.7 | GO:0070458 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.7 | 2.1 | GO:0015920 | regulation of phosphatidylcholine catabolic process(GO:0010899) lipopolysaccharide transport(GO:0015920) |
| 0.6 | 1.7 | GO:0014877 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.4 | 2.6 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.3 | 1.9 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.3 | 3.1 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.3 | 17.4 | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway(GO:0042058) |
| 0.3 | 1.3 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.2 | 1.6 | GO:0000467 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.2 | 0.6 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.1 | 10.1 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 2.1 | GO:0048715 | negative regulation of oligodendrocyte differentiation(GO:0048715) |
| 0.1 | 3.6 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.1 | 2.0 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.1 | 6.8 | GO:0048477 | oogenesis(GO:0048477) |
| 0.1 | 2.6 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.1 | 0.2 | GO:0030862 | regulation of polarized epithelial cell differentiation(GO:0030860) positive regulation of polarized epithelial cell differentiation(GO:0030862) |
| 0.1 | 0.2 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 0.9 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 1.8 | GO:0051569 | regulation of histone H3-K4 methylation(GO:0051569) |
| 0.0 | 1.9 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.3 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 4.9 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.0 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 2.7 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.5 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.3 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.0 | 0.3 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 2.1 | GO:0000082 | G1/S transition of mitotic cell cycle(GO:0000082) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 29.0 | GO:0043218 | compact myelin(GO:0043218) |
| 0.9 | 2.6 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.6 | 3.6 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.4 | 5.7 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.2 | 2.6 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 2.1 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 3.2 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.4 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 8.6 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 17.3 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.1 | 25.2 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 0.3 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 1.3 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 1.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 10.9 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 1.9 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 2.7 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 0.9 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 6.2 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.1 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.6 | GO:0005811 | lipid particle(GO:0005811) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.6 | 3.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.6 | 8.6 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.5 | 29.3 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.4 | 2.2 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.4 | 7.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.3 | 2.6 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.3 | 2.6 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.2 | 17.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.2 | 5.7 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.1 | 6.8 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 3.1 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 5.6 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 2.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 52.2 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 4.7 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 0.4 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.6 | GO:0008748 | N-ethylmaleimide reductase activity(GO:0008748) reduced coenzyme F420 dehydrogenase activity(GO:0043738) sulfur oxygenase reductase activity(GO:0043826) malolactic enzyme activity(GO:0043883) NADPH:sulfur oxidoreductase activity(GO:0043914) epoxyqueuosine reductase activity(GO:0052693) |
| 0.0 | 0.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 1.8 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.9 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |