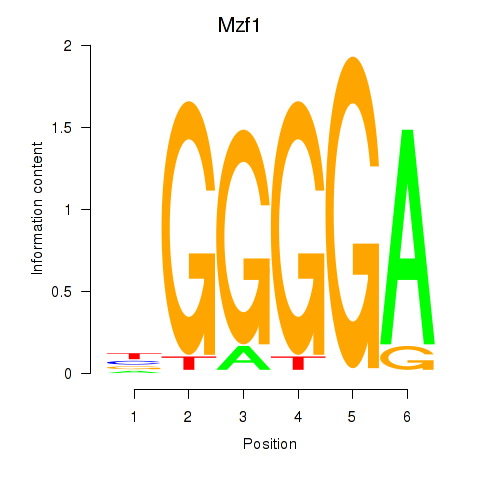
| 2.1 |
6.3 |
GO:0060478 |
acrosomal vesicle exocytosis(GO:0060478) |
| 1.4 |
4.3 |
GO:0097623 |
potassium ion export across plasma membrane(GO:0097623) |
| 1.2 |
3.5 |
GO:0045014 |
detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 1.0 |
2.9 |
GO:0003219 |
cardiac right ventricle formation(GO:0003219) |
| 0.8 |
4.1 |
GO:0097475 |
motor neuron migration(GO:0097475) spinal cord motor neuron migration(GO:0097476) |
| 0.8 |
2.3 |
GO:0017055 |
negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.7 |
2.0 |
GO:0021649 |
vestibulocochlear nerve morphogenesis(GO:0021648) vestibulocochlear nerve structural organization(GO:0021649) ganglion morphogenesis(GO:0061552) dorsal root ganglion morphogenesis(GO:1904835) |
| 0.6 |
1.9 |
GO:0050915 |
sensory perception of sour taste(GO:0050915) |
| 0.6 |
9.8 |
GO:2000821 |
regulation of grooming behavior(GO:2000821) |
| 0.5 |
4.4 |
GO:0048625 |
myoblast fate commitment(GO:0048625) |
| 0.5 |
1.6 |
GO:0035799 |
ureter maturation(GO:0035799) cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) |
| 0.5 |
4.1 |
GO:0035726 |
common myeloid progenitor cell proliferation(GO:0035726) |
| 0.5 |
2.0 |
GO:0002175 |
protein localization to paranode region of axon(GO:0002175) |
| 0.5 |
1.5 |
GO:0006680 |
glucosylceramide catabolic process(GO:0006680) |
| 0.5 |
4.0 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.5 |
0.5 |
GO:1904058 |
positive regulation of sensory perception of pain(GO:1904058) |
| 0.5 |
2.3 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |
| 0.4 |
1.7 |
GO:2000304 |
positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.4 |
4.2 |
GO:0048149 |
behavioral response to ethanol(GO:0048149) |
| 0.4 |
1.2 |
GO:0070428 |
regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.4 |
3.3 |
GO:0061368 |
behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.4 |
0.4 |
GO:1901843 |
positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.4 |
1.6 |
GO:0090238 |
positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.4 |
1.6 |
GO:0021564 |
glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 0.4 |
1.2 |
GO:0002159 |
desmosome assembly(GO:0002159) |
| 0.4 |
1.1 |
GO:0035435 |
phosphate ion transmembrane transport(GO:0035435) |
| 0.4 |
1.4 |
GO:0002074 |
extraocular skeletal muscle development(GO:0002074) pulmonary myocardium development(GO:0003350) subthalamus development(GO:0021539) subthalamic nucleus development(GO:0021763) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) |
| 0.4 |
1.8 |
GO:0030643 |
cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.3 |
1.0 |
GO:0033159 |
negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.3 |
1.0 |
GO:0010643 |
cell communication by chemical coupling(GO:0010643) |
| 0.3 |
1.3 |
GO:0014043 |
negative regulation of neuron maturation(GO:0014043) |
| 0.3 |
1.0 |
GO:1904059 |
regulation of locomotor rhythm(GO:1904059) |
| 0.3 |
0.6 |
GO:0021557 |
oculomotor nerve development(GO:0021557) |
| 0.3 |
1.3 |
GO:0007228 |
positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.3 |
1.0 |
GO:0003096 |
renal sodium ion transport(GO:0003096) |
| 0.3 |
0.9 |
GO:0030070 |
insulin processing(GO:0030070) |
| 0.3 |
1.8 |
GO:0060040 |
retinal bipolar neuron differentiation(GO:0060040) |
| 0.3 |
0.9 |
GO:0032430 |
positive regulation of phospholipase A2 activity(GO:0032430) activation of meiosis involved in egg activation(GO:0060466) |
| 0.3 |
0.6 |
GO:0030240 |
skeletal muscle thin filament assembly(GO:0030240) |
| 0.3 |
0.9 |
GO:0060023 |
soft palate development(GO:0060023) |
| 0.3 |
1.4 |
GO:0015671 |
oxygen transport(GO:0015671) |
| 0.3 |
2.8 |
GO:0048742 |
regulation of skeletal muscle fiber development(GO:0048742) |
| 0.3 |
0.6 |
GO:0070212 |
protein poly-ADP-ribosylation(GO:0070212) |
| 0.3 |
0.8 |
GO:1901187 |
regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.3 |
0.8 |
GO:0071544 |
diphosphoinositol polyphosphate metabolic process(GO:0071543) diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.3 |
1.4 |
GO:2000587 |
regulation of phospholipase A2 activity(GO:0032429) regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.3 |
0.8 |
GO:0030827 |
negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.3 |
0.8 |
GO:1902219 |
regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.3 |
2.3 |
GO:0071420 |
cellular response to histamine(GO:0071420) |
| 0.3 |
0.3 |
GO:0014735 |
regulation of muscle atrophy(GO:0014735) |
| 0.3 |
1.3 |
GO:1902683 |
regulation of receptor localization to synapse(GO:1902683) |
| 0.3 |
1.3 |
GO:0043314 |
negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.3 |
1.8 |
GO:0021555 |
midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.2 |
0.7 |
GO:0060010 |
Sertoli cell fate commitment(GO:0060010) |
| 0.2 |
1.9 |
GO:0036506 |
maintenance of unfolded protein(GO:0036506) protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.2 |
0.2 |
GO:0032241 |
positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.2 |
1.2 |
GO:0090273 |
regulation of somatostatin secretion(GO:0090273) |
| 0.2 |
4.3 |
GO:0021860 |
pyramidal neuron development(GO:0021860) |
| 0.2 |
2.6 |
GO:1901250 |
regulation of lung goblet cell differentiation(GO:1901249) negative regulation of lung goblet cell differentiation(GO:1901250) |
| 0.2 |
1.2 |
GO:0071638 |
negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.2 |
1.4 |
GO:0006177 |
GMP biosynthetic process(GO:0006177) |
| 0.2 |
0.7 |
GO:1990034 |
calcium ion export from cell(GO:1990034) |
| 0.2 |
0.7 |
GO:0044339 |
canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.2 |
0.2 |
GO:0060591 |
chondroblast differentiation(GO:0060591) |
| 0.2 |
3.1 |
GO:0071625 |
vocalization behavior(GO:0071625) |
| 0.2 |
1.5 |
GO:0042699 |
follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.2 |
4.4 |
GO:1903861 |
regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.2 |
1.1 |
GO:0002121 |
inter-male aggressive behavior(GO:0002121) |
| 0.2 |
1.2 |
GO:0072318 |
clathrin coat disassembly(GO:0072318) |
| 0.2 |
2.2 |
GO:0010650 |
positive regulation of cell communication by electrical coupling(GO:0010650) regulation of membrane depolarization during action potential(GO:0098902) regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 |
0.8 |
GO:2000774 |
positive regulation of cellular senescence(GO:2000774) |
| 0.2 |
1.2 |
GO:1903056 |
regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.2 |
1.4 |
GO:1900194 |
negative regulation of oocyte maturation(GO:1900194) |
| 0.2 |
2.2 |
GO:1902018 |
negative regulation of cilium assembly(GO:1902018) |
| 0.2 |
1.0 |
GO:0031915 |
positive regulation of synaptic plasticity(GO:0031915) |
| 0.2 |
1.2 |
GO:0060024 |
rhythmic synaptic transmission(GO:0060024) |
| 0.2 |
0.6 |
GO:1903244 |
positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.2 |
0.8 |
GO:0051964 |
negative regulation of synapse assembly(GO:0051964) |
| 0.2 |
0.8 |
GO:1904177 |
regulation of adipose tissue development(GO:1904177) |
| 0.2 |
1.1 |
GO:0060836 |
lymphatic endothelial cell differentiation(GO:0060836) |
| 0.2 |
1.5 |
GO:0000395 |
mRNA 5'-splice site recognition(GO:0000395) |
| 0.2 |
1.2 |
GO:0036072 |
intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.2 |
0.7 |
GO:0019919 |
peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.2 |
0.7 |
GO:0051562 |
negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.2 |
0.5 |
GO:0038003 |
opioid receptor signaling pathway(GO:0038003) regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.2 |
1.2 |
GO:0007195 |
adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.2 |
0.8 |
GO:0071787 |
endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.2 |
1.0 |
GO:0001831 |
trophectodermal cellular morphogenesis(GO:0001831) |
| 0.2 |
0.5 |
GO:0070459 |
prolactin secretion(GO:0070459) |
| 0.2 |
1.6 |
GO:0045161 |
neuronal ion channel clustering(GO:0045161) |
| 0.2 |
2.1 |
GO:0060081 |
membrane hyperpolarization(GO:0060081) |
| 0.2 |
1.5 |
GO:0033574 |
response to testosterone(GO:0033574) |
| 0.2 |
1.2 |
GO:0097501 |
stress response to metal ion(GO:0097501) |
| 0.1 |
0.6 |
GO:0051311 |
meiotic metaphase plate congression(GO:0051311) |
| 0.1 |
0.9 |
GO:0021814 |
cell motility involved in cerebral cortex radial glia guided migration(GO:0021814) |
| 0.1 |
0.9 |
GO:0072321 |
chaperone-mediated protein transport(GO:0072321) |
| 0.1 |
0.7 |
GO:0035262 |
gonad morphogenesis(GO:0035262) |
| 0.1 |
0.8 |
GO:0090267 |
positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.1 |
1.2 |
GO:0098907 |
protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) regulation of atrial cardiac muscle cell action potential(GO:0098910) |
| 0.1 |
1.2 |
GO:0045742 |
positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.1 |
1.4 |
GO:0071225 |
cellular response to muramyl dipeptide(GO:0071225) |
| 0.1 |
0.7 |
GO:0098828 |
modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.1 |
1.6 |
GO:0042048 |
olfactory behavior(GO:0042048) |
| 0.1 |
0.3 |
GO:0071550 |
death-inducing signaling complex assembly(GO:0071550) |
| 0.1 |
1.9 |
GO:0031507 |
heterochromatin assembly(GO:0031507) |
| 0.1 |
1.6 |
GO:0061577 |
calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) |
| 0.1 |
0.6 |
GO:1905098 |
negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.1 |
0.4 |
GO:0001866 |
NK T cell proliferation(GO:0001866) |
| 0.1 |
1.7 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 |
1.1 |
GO:0045974 |
miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 |
1.2 |
GO:0030322 |
stabilization of membrane potential(GO:0030322) |
| 0.1 |
1.4 |
GO:2001223 |
negative regulation of neuron migration(GO:2001223) |
| 0.1 |
0.8 |
GO:0034047 |
regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.1 |
2.0 |
GO:0016486 |
peptide hormone processing(GO:0016486) |
| 0.1 |
1.9 |
GO:0002091 |
negative regulation of receptor internalization(GO:0002091) |
| 0.1 |
0.7 |
GO:0021797 |
forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 |
0.8 |
GO:0030263 |
apoptotic chromosome condensation(GO:0030263) |
| 0.1 |
0.4 |
GO:0030576 |
Cajal body organization(GO:0030576) |
| 0.1 |
1.0 |
GO:0060013 |
righting reflex(GO:0060013) positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.1 |
1.3 |
GO:0048672 |
positive regulation of collateral sprouting(GO:0048672) |
| 0.1 |
0.5 |
GO:0002182 |
cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.1 |
0.6 |
GO:0051798 |
positive regulation of hair follicle development(GO:0051798) |
| 0.1 |
0.4 |
GO:1900122 |
positive regulation of receptor binding(GO:1900122) |
| 0.1 |
0.3 |
GO:0036166 |
phenotypic switching(GO:0036166) |
| 0.1 |
0.5 |
GO:0032811 |
negative regulation of epinephrine secretion(GO:0032811) |
| 0.1 |
0.4 |
GO:0007023 |
post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 |
2.0 |
GO:0048305 |
immunoglobulin secretion(GO:0048305) |
| 0.1 |
1.1 |
GO:0033148 |
positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.1 |
0.3 |
GO:0086013 |
membrane repolarization during cardiac muscle cell action potential(GO:0086013) |
| 0.1 |
0.2 |
GO:0010726 |
positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.1 |
1.0 |
GO:0007379 |
segment specification(GO:0007379) |
| 0.1 |
0.4 |
GO:0035624 |
receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.1 |
1.0 |
GO:0032482 |
Rab protein signal transduction(GO:0032482) |
| 0.1 |
0.7 |
GO:0023035 |
CD40 signaling pathway(GO:0023035) |
| 0.1 |
0.4 |
GO:1903966 |
monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 |
0.2 |
GO:1904457 |
positive regulation of neuronal action potential(GO:1904457) |
| 0.1 |
0.7 |
GO:2000786 |
positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 |
0.3 |
GO:0019405 |
alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 |
0.4 |
GO:0046898 |
response to cycloheximide(GO:0046898) |
| 0.1 |
0.3 |
GO:0048162 |
preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.1 |
1.1 |
GO:0050966 |
detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.1 |
0.3 |
GO:0097118 |
neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.1 |
0.3 |
GO:0065001 |
negative regulation of histone H3-K36 methylation(GO:0000415) specification of axis polarity(GO:0065001) |
| 0.1 |
2.4 |
GO:0097320 |
membrane tubulation(GO:0097320) |
| 0.1 |
1.6 |
GO:0034063 |
stress granule assembly(GO:0034063) |
| 0.1 |
0.9 |
GO:0007252 |
I-kappaB phosphorylation(GO:0007252) |
| 0.1 |
0.2 |
GO:0001543 |
ovarian follicle rupture(GO:0001543) |
| 0.1 |
1.5 |
GO:0045063 |
T-helper 1 cell differentiation(GO:0045063) |
| 0.1 |
1.2 |
GO:1990403 |
embryonic brain development(GO:1990403) |
| 0.1 |
0.8 |
GO:0060159 |
regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.1 |
0.2 |
GO:0002069 |
columnar/cuboidal epithelial cell maturation(GO:0002069) |
| 0.1 |
2.3 |
GO:0061098 |
positive regulation of protein tyrosine kinase activity(GO:0061098) |
| 0.1 |
0.4 |
GO:0010991 |
regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 |
0.4 |
GO:0040032 |
post-embryonic body morphogenesis(GO:0040032) |
| 0.1 |
1.6 |
GO:2001222 |
regulation of neuron migration(GO:2001222) |
| 0.1 |
0.3 |
GO:0030432 |
peristalsis(GO:0030432) |
| 0.1 |
3.6 |
GO:0007019 |
microtubule depolymerization(GO:0007019) |
| 0.1 |
0.8 |
GO:0051601 |
exocyst localization(GO:0051601) |
| 0.1 |
0.3 |
GO:0060368 |
regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 0.1 |
1.1 |
GO:0042541 |
hemoglobin biosynthetic process(GO:0042541) |
| 0.1 |
0.8 |
GO:2001241 |
positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.1 |
0.2 |
GO:0090135 |
actin filament branching(GO:0090135) negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.1 |
1.3 |
GO:0031115 |
negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 |
0.2 |
GO:0070315 |
G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.1 |
0.2 |
GO:1901978 |
positive regulation of cell cycle checkpoint(GO:1901978) |
| 0.1 |
0.8 |
GO:0043252 |
oligopeptide transport(GO:0006857) prostaglandin transport(GO:0015732) sodium-independent organic anion transport(GO:0043252) |
| 0.1 |
0.6 |
GO:0021869 |
forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 |
0.2 |
GO:1904274 |
tricellular tight junction assembly(GO:1904274) |
| 0.1 |
0.1 |
GO:0021698 |
cerebellar cortex structural organization(GO:0021698) |
| 0.1 |
0.7 |
GO:0021854 |
hypothalamus development(GO:0021854) |
| 0.1 |
1.0 |
GO:0021902 |
commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 |
0.6 |
GO:0035970 |
peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 |
0.3 |
GO:0098734 |
protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 |
1.0 |
GO:0080182 |
histone H3-K4 trimethylation(GO:0080182) |
| 0.1 |
0.8 |
GO:0043982 |
histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 |
0.2 |
GO:0006438 |
valyl-tRNA aminoacylation(GO:0006438) |
| 0.1 |
0.8 |
GO:0009072 |
aromatic amino acid family metabolic process(GO:0009072) |
| 0.1 |
0.2 |
GO:0072186 |
canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) lactic acid secretion(GO:0046722) metanephric cap development(GO:0072185) metanephric cap morphogenesis(GO:0072186) metanephric cap mesenchymal cell proliferation involved in metanephros development(GO:0090094) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.1 |
0.4 |
GO:0030854 |
positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 |
2.8 |
GO:0046513 |
ceramide biosynthetic process(GO:0046513) |
| 0.1 |
1.4 |
GO:0048843 |
negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.1 |
0.7 |
GO:0048280 |
vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 |
0.1 |
GO:0002741 |
positive regulation of cytokine secretion involved in immune response(GO:0002741) |
| 0.1 |
0.2 |
GO:0006421 |
asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 |
0.3 |
GO:0006384 |
transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 |
0.4 |
GO:0018230 |
peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 |
0.1 |
GO:0021506 |
anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.1 |
1.1 |
GO:0043171 |
peptide catabolic process(GO:0043171) |
| 0.1 |
1.1 |
GO:0048268 |
clathrin coat assembly(GO:0048268) |
| 0.1 |
0.4 |
GO:0097119 |
postsynaptic density protein 95 clustering(GO:0097119) |
| 0.1 |
0.2 |
GO:0035928 |
rRNA import into mitochondrion(GO:0035928) |
| 0.0 |
1.0 |
GO:0097120 |
receptor localization to synapse(GO:0097120) |
| 0.0 |
0.3 |
GO:0006362 |
transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 |
0.8 |
GO:0060384 |
innervation(GO:0060384) |
| 0.0 |
0.5 |
GO:0021942 |
radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 |
0.4 |
GO:0075522 |
IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 |
0.3 |
GO:0071243 |
cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 |
0.2 |
GO:0021914 |
negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 |
1.0 |
GO:0048147 |
negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 |
0.4 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 |
0.2 |
GO:0090383 |
phagosome acidification(GO:0090383) |
| 0.0 |
0.2 |
GO:0036515 |
serotonergic neuron axon guidance(GO:0036515) |
| 0.0 |
0.9 |
GO:2000785 |
regulation of autophagosome assembly(GO:2000785) |
| 0.0 |
0.2 |
GO:0031642 |
negative regulation of myelination(GO:0031642) negative regulation of translation in response to stress(GO:0032055) |
| 0.0 |
1.4 |
GO:0060135 |
maternal process involved in female pregnancy(GO:0060135) |
| 0.0 |
0.9 |
GO:0030901 |
midbrain development(GO:0030901) |
| 0.0 |
0.9 |
GO:0010800 |
positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 |
0.5 |
GO:0051382 |
kinetochore assembly(GO:0051382) |
| 0.0 |
0.0 |
GO:0030222 |
eosinophil differentiation(GO:0030222) |
| 0.0 |
0.4 |
GO:1901223 |
negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 |
0.6 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 0.0 |
0.6 |
GO:0035024 |
negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 |
0.6 |
GO:0045717 |
negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 |
0.3 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.0 |
0.9 |
GO:0071806 |
intracellular protein transmembrane transport(GO:0065002) protein transmembrane transport(GO:0071806) |
| 0.0 |
0.2 |
GO:1904690 |
regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.0 |
0.2 |
GO:0046339 |
diacylglycerol metabolic process(GO:0046339) |
| 0.0 |
0.1 |
GO:2000501 |
natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.0 |
0.1 |
GO:0045213 |
neurotransmitter receptor metabolic process(GO:0045213) |
| 0.0 |
0.1 |
GO:0045925 |
positive regulation of female receptivity(GO:0045925) |
| 0.0 |
0.6 |
GO:0043486 |
histone exchange(GO:0043486) |
| 0.0 |
0.1 |
GO:0070649 |
meiotic chromosome movement towards spindle pole(GO:0016344) formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 |
0.4 |
GO:1900452 |
regulation of long term synaptic depression(GO:1900452) |
| 0.0 |
0.2 |
GO:0043984 |
histone H4-K16 acetylation(GO:0043984) |
| 0.0 |
1.2 |
GO:0000381 |
regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 |
0.0 |
GO:1901097 |
negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 |
0.7 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.0 |
0.1 |
GO:1903300 |
negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 |
0.2 |
GO:2000601 |
positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 |
0.2 |
GO:0033564 |
anterior/posterior axon guidance(GO:0033564) |
| 0.0 |
0.3 |
GO:2001256 |
regulation of store-operated calcium entry(GO:2001256) |
| 0.0 |
2.1 |
GO:0001578 |
microtubule bundle formation(GO:0001578) |
| 0.0 |
0.1 |
GO:0070889 |
platelet alpha granule organization(GO:0070889) |
| 0.0 |
0.3 |
GO:0032785 |
negative regulation of DNA-templated transcription, elongation(GO:0032785) |
| 0.0 |
0.6 |
GO:0043967 |
histone H4 acetylation(GO:0043967) |
| 0.0 |
0.2 |
GO:1904707 |
positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.0 |
0.2 |
GO:0044375 |
regulation of peroxisome size(GO:0044375) |
| 0.0 |
0.4 |
GO:0071391 |
cellular response to estrogen stimulus(GO:0071391) |
| 0.0 |
0.9 |
GO:0007029 |
endoplasmic reticulum organization(GO:0007029) |
| 0.0 |
0.1 |
GO:0034316 |
negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 |
0.4 |
GO:0006353 |
DNA-templated transcription, termination(GO:0006353) |
| 0.0 |
0.7 |
GO:0051865 |
protein autoubiquitination(GO:0051865) |
| 0.0 |
0.7 |
GO:0031061 |
negative regulation of histone methylation(GO:0031061) |
| 0.0 |
1.6 |
GO:0051965 |
positive regulation of synapse assembly(GO:0051965) |
| 0.0 |
0.1 |
GO:0035542 |
regulation of SNARE complex assembly(GO:0035542) |
| 0.0 |
0.5 |
GO:1901385 |
regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 |
0.2 |
GO:0070935 |
3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 |
0.2 |
GO:0034122 |
negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.0 |
0.2 |
GO:0048026 |
positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 |
0.2 |
GO:0010839 |
negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 |
0.5 |
GO:0006625 |
protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 |
1.2 |
GO:0070936 |
protein K48-linked ubiquitination(GO:0070936) |
| 0.0 |
0.1 |
GO:0072592 |
oxygen metabolic process(GO:0072592) |
| 0.0 |
0.4 |
GO:0016239 |
positive regulation of macroautophagy(GO:0016239) positive regulation of response to extracellular stimulus(GO:0032106) positive regulation of response to nutrient levels(GO:0032109) |
| 0.0 |
0.3 |
GO:0060218 |
hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 |
0.4 |
GO:0051123 |
RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 |
0.1 |
GO:0002922 |
positive regulation of humoral immune response(GO:0002922) |
| 0.0 |
0.3 |
GO:0031167 |
rRNA methylation(GO:0031167) |
| 0.0 |
2.3 |
GO:0000209 |
protein polyubiquitination(GO:0000209) |
| 0.0 |
0.4 |
GO:0070208 |
protein heterotrimerization(GO:0070208) |
| 0.0 |
0.2 |
GO:0000042 |
protein targeting to Golgi(GO:0000042) |
| 0.0 |
0.2 |
GO:0046007 |
negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 |
0.3 |
GO:0006744 |
ubiquinone biosynthetic process(GO:0006744) |
| 0.0 |
0.2 |
GO:0048172 |
regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 |
1.1 |
GO:0010923 |
negative regulation of phosphatase activity(GO:0010923) |
| 0.0 |
0.2 |
GO:0000154 |
rRNA modification(GO:0000154) |
| 0.0 |
0.1 |
GO:0072531 |
pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.0 |
0.1 |
GO:0034227 |
tRNA thio-modification(GO:0034227) |
| 0.0 |
0.1 |
GO:0090666 |
scaRNA localization to Cajal body(GO:0090666) |
| 0.0 |
0.2 |
GO:0048025 |
negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 |
0.7 |
GO:0046579 |
positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.0 |
0.0 |
GO:0061428 |
negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.0 |
0.5 |
GO:0010507 |
negative regulation of autophagy(GO:0010507) |
| 0.0 |
0.1 |
GO:0045199 |
maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 |
0.1 |
GO:0051189 |
Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 |
0.2 |
GO:0008535 |
respiratory chain complex IV assembly(GO:0008535) |
| 0.0 |
0.1 |
GO:0030252 |
growth hormone secretion(GO:0030252) |
| 0.0 |
0.4 |
GO:0016578 |
histone deubiquitination(GO:0016578) |
| 0.0 |
0.2 |
GO:0045793 |
positive regulation of cell size(GO:0045793) |
| 0.0 |
0.3 |
GO:0010569 |
regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 |
0.1 |
GO:0090179 |
planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 |
0.7 |
GO:0043966 |
histone H3 acetylation(GO:0043966) |
| 0.0 |
0.3 |
GO:0010614 |
negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 |
0.7 |
GO:0002028 |
regulation of sodium ion transport(GO:0002028) |
| 0.0 |
0.3 |
GO:0071108 |
protein K48-linked deubiquitination(GO:0071108) |
| 0.0 |
0.4 |
GO:0000184 |
nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 |
0.1 |
GO:0034058 |
endosomal vesicle fusion(GO:0034058) |
| 0.0 |
0.1 |
GO:0006999 |
nuclear pore organization(GO:0006999) |
| 0.0 |
0.3 |
GO:0006879 |
cellular iron ion homeostasis(GO:0006879) |
| 0.0 |
0.1 |
GO:1900271 |
regulation of long-term synaptic potentiation(GO:1900271) |
| 0.0 |
0.2 |
GO:0045116 |
protein neddylation(GO:0045116) |
| 0.0 |
0.3 |
GO:0046580 |
negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 |
0.1 |
GO:0007216 |
G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 |
0.0 |
GO:1903378 |
positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 |
0.1 |
GO:1901621 |
regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901620) negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 |
0.0 |
GO:0018343 |
protein farnesylation(GO:0018343) |
| 0.0 |
0.1 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 |
0.3 |
GO:0006891 |
intra-Golgi vesicle-mediated transport(GO:0006891) |