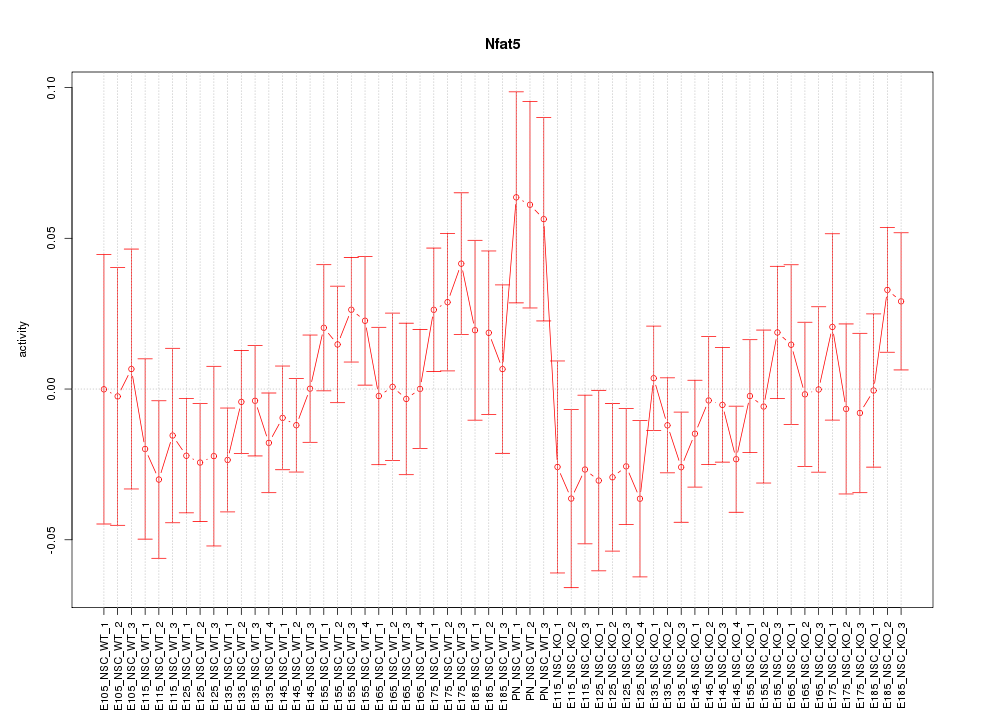
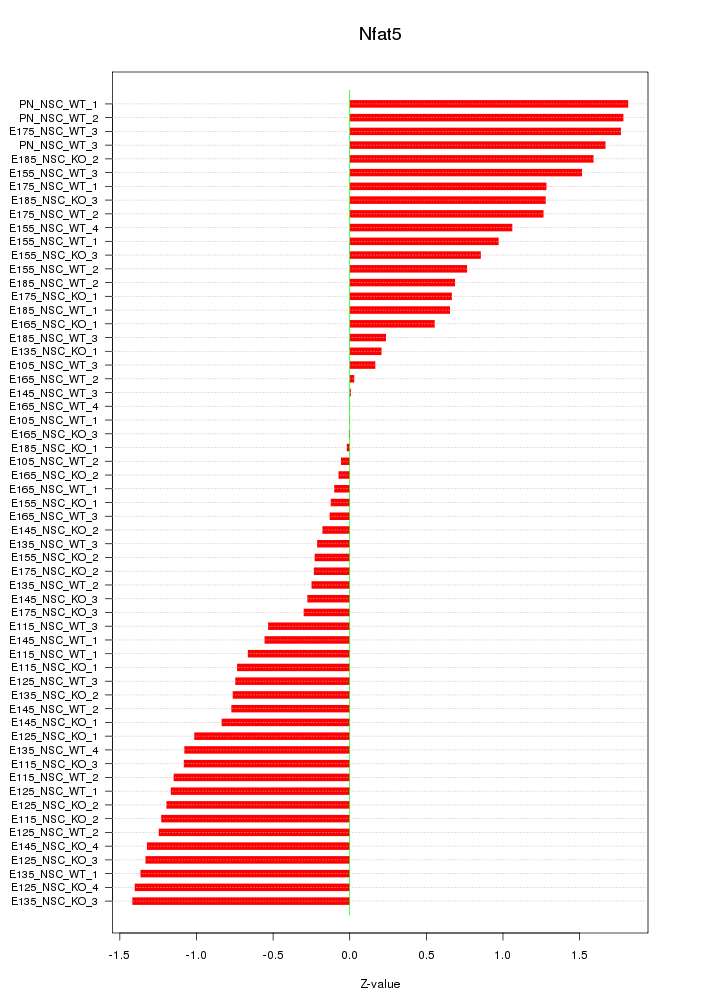
Motif ID: Nfat5
Z-value: 0.936
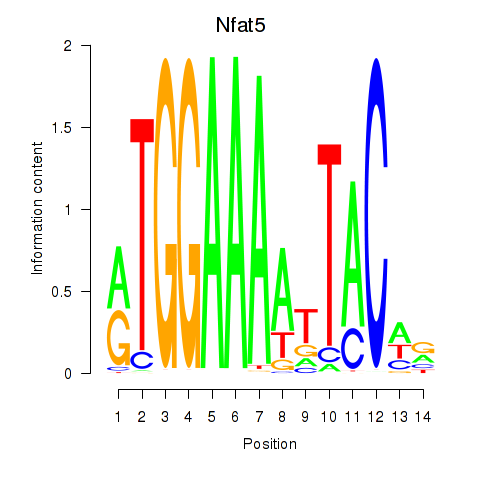
Transcription factors associated with Nfat5:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nfat5 | ENSMUSG00000003847.10 | Nfat5 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfat5 | mm10_v2_chr8_+_107293463_107293483 | 0.20 | 1.3e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.8 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 2.3 | 7.0 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 2.2 | 8.8 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 1.4 | 10.9 | GO:0032264 | IMP salvage(GO:0032264) |
| 1.1 | 3.3 | GO:0003032 | detection of oxygen(GO:0003032) regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905005) |
| 1.1 | 7.4 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) follicle-stimulating hormone signaling pathway(GO:0042699) |
| 1.0 | 3.0 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.9 | 9.4 | GO:0042447 | hormone catabolic process(GO:0042447) |
| 0.8 | 2.5 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.7 | 2.8 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.6 | 1.9 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.5 | 2.0 | GO:0060745 | mammary gland branching involved in pregnancy(GO:0060745) |
| 0.5 | 4.4 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.4 | 5.4 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.4 | 1.9 | GO:0032911 | nerve growth factor production(GO:0032902) negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.3 | 2.4 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.3 | 9.8 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.3 | 1.8 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.3 | 1.9 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.3 | 1.0 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.2 | 0.7 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.2 | 1.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.2 | 4.8 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.2 | 3.3 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.2 | 3.1 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.2 | 1.1 | GO:1903352 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.1 | 1.3 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 1.0 | GO:0015791 | polyol transport(GO:0015791) |
| 0.1 | 1.1 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.1 | 1.7 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 1.0 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.1 | 1.3 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 0.5 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.1 | 4.7 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.1 | 4.4 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.1 | 11.0 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.1 | 6.1 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.5 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 2.8 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 0.7 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 1.9 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 3.2 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.0 | 0.9 | GO:0090630 | activation of GTPase activity(GO:0090630) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 7.0 | GO:0044327 | dendritic spine membrane(GO:0032591) dendritic spine head(GO:0044327) |
| 0.5 | 7.4 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.4 | 8.8 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.4 | 3.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.2 | 4.8 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.2 | 1.0 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 1.9 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 9.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 1.9 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 2.0 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 0.7 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 1.2 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.1 | 4.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 3.3 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 1.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 4.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.7 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 2.5 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.0 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 2.8 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 8.5 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 18.7 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 18.0 | GO:0005829 | cytosol(GO:0005829) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.8 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 2.5 | 7.4 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 1.6 | 10.9 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 1.5 | 9.0 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 1.3 | 9.4 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 1.1 | 4.4 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.9 | 7.0 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.8 | 3.3 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.8 | 3.1 | GO:0086038 | calcium:sodium antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086038) |
| 0.8 | 3.0 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.7 | 4.8 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.7 | 5.4 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.5 | 11.0 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.5 | 1.9 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.4 | 1.8 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.3 | 1.0 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.3 | 1.9 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.3 | 8.8 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.2 | 2.0 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.2 | 1.9 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.2 | 1.3 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.2 | 2.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 1.1 | GO:0015189 | L-ornithine transmembrane transporter activity(GO:0000064) arginine transmembrane transporter activity(GO:0015181) L-lysine transmembrane transporter activity(GO:0015189) |
| 0.2 | 0.5 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.1 | 1.7 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.1 | 1.0 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.1 | 1.2 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 6.1 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 5.2 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.7 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 2.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 4.6 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.7 | GO:0005518 | collagen binding(GO:0005518) |