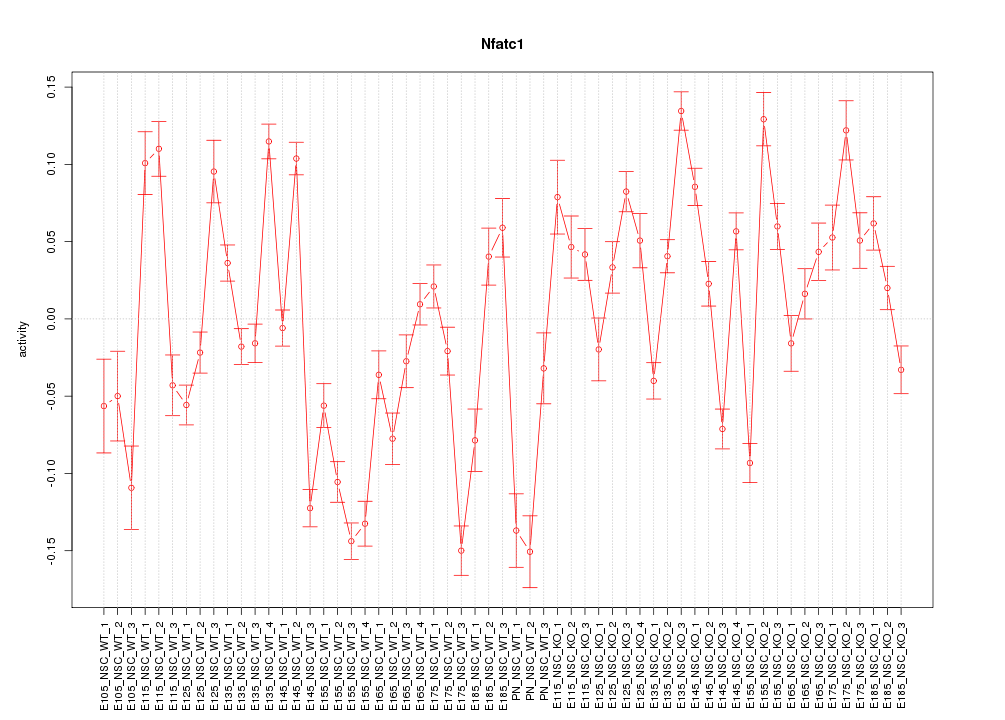
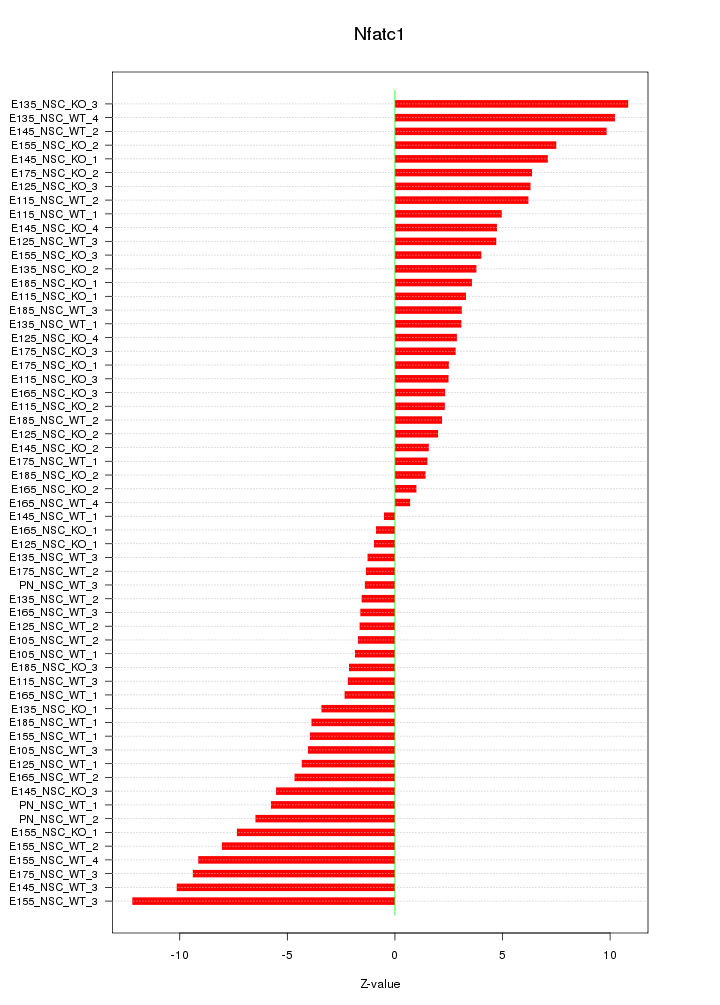
Motif ID: Nfatc1
Z-value: 5.087
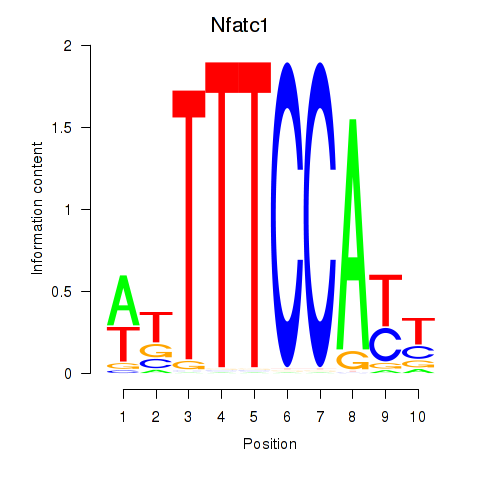
Transcription factors associated with Nfatc1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nfatc1 | ENSMUSG00000033016.9 | Nfatc1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfatc1 | mm10_v2_chr18_-_80713062_80713080 | -0.33 | 1.1e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 14.7 | GO:0003345 | proepicardium cell migration involved in pericardium morphogenesis(GO:0003345) |
| 2.9 | 20.5 | GO:0072539 | T-helper 17 cell differentiation(GO:0072539) |
| 2.8 | 5.7 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 2.4 | 14.2 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 1.7 | 6.9 | GO:0035795 | negative regulation of mitochondrial membrane permeability(GO:0035795) |
| 1.5 | 4.4 | GO:0060126 | somatotropin secreting cell differentiation(GO:0060126) |
| 1.4 | 4.3 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 1.4 | 5.6 | GO:0097393 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 1.4 | 5.5 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 1.2 | 7.2 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 1.1 | 8.9 | GO:0046645 | positive regulation of gamma-delta T cell differentiation(GO:0045588) positive regulation of gamma-delta T cell activation(GO:0046645) positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.9 | 11.4 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.9 | 3.5 | GO:0032439 | endosome localization(GO:0032439) |
| 0.8 | 2.3 | GO:0060084 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) circadian sleep/wake cycle, REM sleep(GO:0042747) synaptic transmission involved in micturition(GO:0060084) |
| 0.7 | 7.2 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.6 | 3.6 | GO:0051461 | regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.6 | 2.3 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.6 | 4.0 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.6 | 1.7 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.5 | 2.7 | GO:0019659 | lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.5 | 1.5 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.5 | 14.9 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.4 | 2.6 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.3 | 5.2 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.3 | 1.8 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.3 | 5.1 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.3 | 1.4 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.3 | 3.0 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.3 | 1.3 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.2 | 1.4 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.2 | 3.3 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.2 | 0.9 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.2 | 2.3 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.2 | 0.7 | GO:0090135 | actin filament branching(GO:0090135) negative regulation of phospholipase C activity(GO:1900275) |
| 0.2 | 0.4 | GO:0001787 | natural killer cell proliferation(GO:0001787) |
| 0.2 | 2.3 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.2 | 1.9 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.2 | 1.7 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.2 | 2.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.2 | 5.1 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.2 | 1.1 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.2 | 3.4 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.2 | 6.6 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.1 | 1.4 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 1.3 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 0.1 | 1.3 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.1 | 1.4 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 0.3 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.1 | 0.6 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 5.0 | GO:0042491 | auditory receptor cell differentiation(GO:0042491) |
| 0.1 | 0.3 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.1 | 0.7 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.1 | 0.2 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.1 | 2.1 | GO:0032148 | activation of protein kinase B activity(GO:0032148) |
| 0.1 | 2.9 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.1 | 8.5 | GO:0006497 | protein lipidation(GO:0006497) |
| 0.1 | 0.4 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 0.7 | GO:0007379 | segment specification(GO:0007379) |
| 0.1 | 1.1 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 0.4 | GO:0072488 | nitrogen utilization(GO:0019740) ammonium transmembrane transport(GO:0072488) |
| 0.1 | 1.3 | GO:0010640 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) |
| 0.1 | 1.3 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.1 | 1.1 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.1 | 0.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.1 | GO:0061218 | negative regulation of mesonephros development(GO:0061218) negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 0.1 | 1230.1 | GO:0008150 | biological_process(GO:0008150) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 7.2 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 1.4 | 5.6 | GO:1990421 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 0.9 | 3.6 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.8 | 3.4 | GO:0031673 | H zone(GO:0031673) |
| 0.6 | 3.5 | GO:0045179 | apical cortex(GO:0045179) |
| 0.4 | 8.9 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.3 | 14.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.3 | 5.8 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.2 | 0.7 | GO:0000811 | GINS complex(GO:0000811) |
| 0.2 | 2.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 2.7 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 3.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 4.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 3.5 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 1.8 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 1362.1 | GO:0005575 | cellular_component(GO:0005575) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 20.5 | GO:0008142 | oxysterol binding(GO:0008142) |
| 2.5 | 7.4 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 1.8 | 8.9 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 1.4 | 6.9 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.8 | 5.6 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.7 | 3.6 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.7 | 2.7 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.7 | 14.7 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.6 | 2.5 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.5 | 1.5 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.5 | 1.4 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.4 | 3.0 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.4 | 1.4 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.3 | 7.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.3 | 14.2 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.3 | 3.9 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.3 | 0.8 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.3 | 8.2 | GO:0034930 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) heparan sulfate 2-O-sulfotransferase activity(GO:0004394) HNK-1 sulfotransferase activity(GO:0016232) heparan sulfate 6-O-sulfotransferase activity(GO:0017095) trans-9R,10R-dihydrodiolphenanthrene sulfotransferase activity(GO:0018721) 1-phenanthrol sulfotransferase activity(GO:0018722) 3-phenanthrol sulfotransferase activity(GO:0018723) 4-phenanthrol sulfotransferase activity(GO:0018724) trans-3,4-dihydrodiolphenanthrene sulfotransferase activity(GO:0018725) 9-phenanthrol sulfotransferase activity(GO:0018726) 2-phenanthrol sulfotransferase activity(GO:0018727) phenanthrol sulfotransferase activity(GO:0019111) 1-hydroxypyrene sulfotransferase activity(GO:0034930) proteoglycan sulfotransferase activity(GO:0050698) cholesterol sulfotransferase activity(GO:0051922) hydroxyjasmonate sulfotransferase activity(GO:0080131) |
| 0.2 | 3.7 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.2 | 7.8 | GO:0034945 | dihydrolipoamide branched chain acyltransferase activity(GO:0004147) palmitoleoyl [acyl-carrier-protein]-dependent acyltransferase activity(GO:0008951) serine O-acyltransferase activity(GO:0016412) O-succinyltransferase activity(GO:0016750) sinapoyltransferase activity(GO:0016752) O-sinapoyltransferase activity(GO:0016753) peptidyl-lysine N6-myristoyltransferase activity(GO:0018030) peptidyl-lysine N6-palmitoyltransferase activity(GO:0018031) benzoyl acetate-CoA thiolase activity(GO:0018711) 3-hydroxybutyryl-CoA thiolase activity(GO:0018712) 3-ketopimelyl-CoA thiolase activity(GO:0018713) N-palmitoyltransferase activity(GO:0019105) acyl-CoA N-acyltransferase activity(GO:0019186) protein-cysteine S-myristoyltransferase activity(GO:0019705) glucosaminyl-phosphotidylinositol O-acyltransferase activity(GO:0032216) ergosterol O-acyltransferase activity(GO:0034737) lanosterol O-acyltransferase activity(GO:0034738) naphthyl-2-oxomethyl-succinyl-CoA succinyl transferase activity(GO:0034848) 2,4,4-trimethyl-3-oxopentanoyl-CoA 2-C-propanoyl transferase activity(GO:0034851) 2-methylhexanoyl-CoA C-acetyltransferase activity(GO:0034915) butyryl-CoA 2-C-propionyltransferase activity(GO:0034919) 2,6-dimethyl-5-methylene-3-oxo-heptanoyl-CoA C-acetyltransferase activity(GO:0034945) L-2-aminoadipate N-acetyltransferase activity(GO:0043741) keto acid formate lyase activity(GO:0043806) azetidine-2-carboxylic acid acetyltransferase activity(GO:0046941) peptidyl-lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0052858) acetyl-CoA:L-lysine N6-acetyltransferase(GO:0090595) |
| 0.2 | 0.9 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.2 | 6.6 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.2 | 2.3 | GO:0004889 | acetylcholine-activated cation-selective channel activity(GO:0004889) |
| 0.2 | 5.5 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.2 | 11.0 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.2 | 2.3 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 0.8 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.1 | 9.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 0.6 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 2.1 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 1.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.3 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.1 | 12.3 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 1.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 2.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 3.8 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 1.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 2.3 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 1.6 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 1295.5 | GO:0003674 | molecular_function(GO:0003674) |