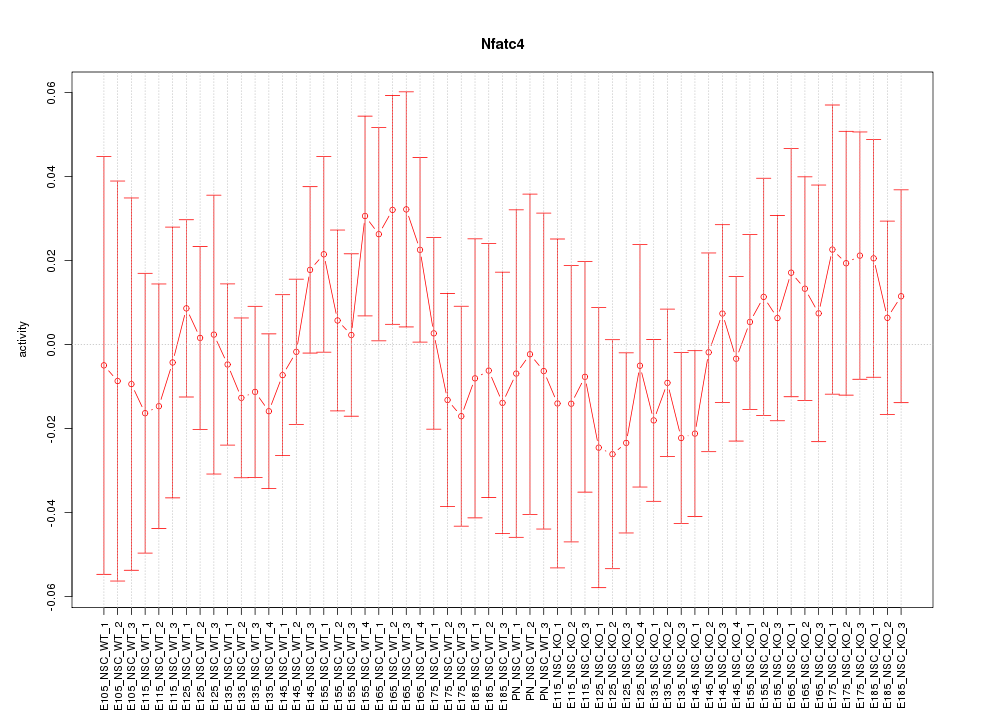
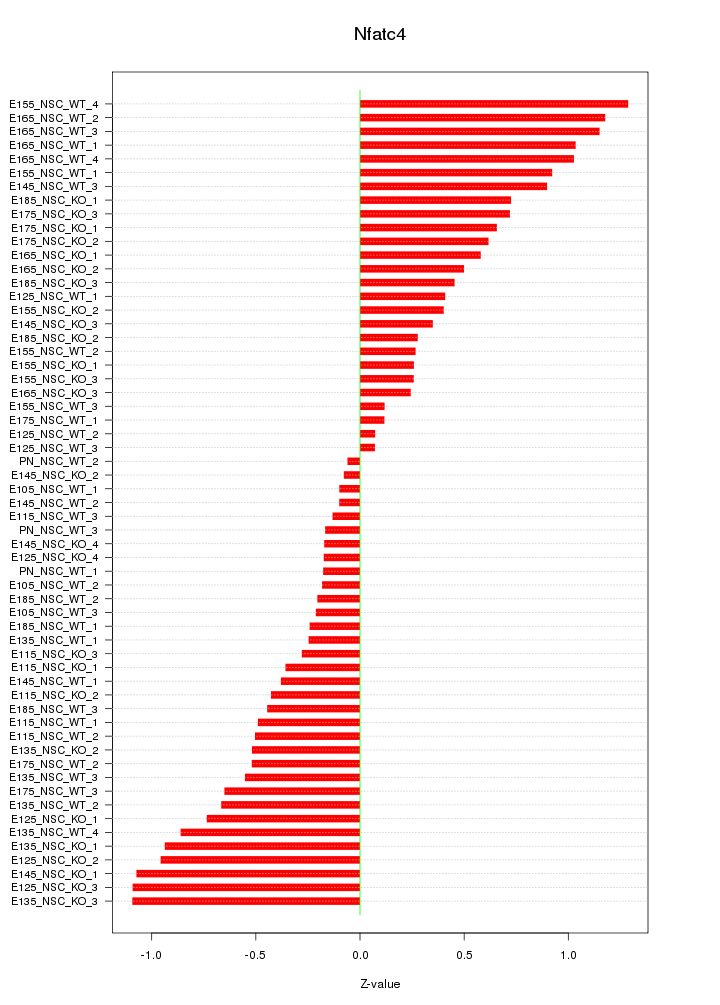
Motif ID: Nfatc4
Z-value: 0.605
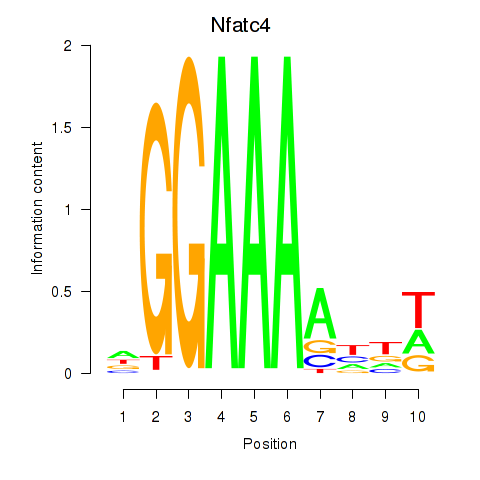
Transcription factors associated with Nfatc4:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nfatc4 | ENSMUSG00000023411.5 | Nfatc4 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfatc4 | mm10_v2_chr14_+_55824795_55824889 | -0.32 | 1.4e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.3 | GO:0038095 | positive regulation of mast cell cytokine production(GO:0032765) Fc-epsilon receptor signaling pathway(GO:0038095) |
| 2.0 | 5.9 | GO:0007521 | muscle cell fate determination(GO:0007521) cellular response to parathyroid hormone stimulus(GO:0071374) positive regulation of macrophage apoptotic process(GO:2000111) |
| 1.2 | 3.6 | GO:0070428 | regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.9 | 13.8 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.6 | 5.4 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.4 | 1.2 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.4 | 2.5 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.3 | 1.0 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.2 | 1.2 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.2 | 2.0 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.2 | 0.5 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.2 | 0.6 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.1 | 0.6 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 2.8 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 0.5 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 1.6 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 1.0 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 1.7 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:1905005 | regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905005) |
| 0.0 | 2.3 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.5 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.4 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.3 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.5 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.7 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.3 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 1.9 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 2.6 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.3 | 5.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.3 | 1.0 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 15.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 2.0 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 1.2 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 5.9 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 2.5 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 2.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.0 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 1.7 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 4.3 | GO:0005667 | transcription factor complex(GO:0005667) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.8 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.5 | 5.9 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.4 | 5.8 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 0.6 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 1.9 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 1.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.9 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 2.0 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.1 | 1.0 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 5.4 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 1.2 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 0.4 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 1.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 1.2 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 2.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 1.6 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.4 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.3 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.5 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 1.7 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 2.2 | GO:0001948 | glycoprotein binding(GO:0001948) |
| 0.0 | 3.6 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 1.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |