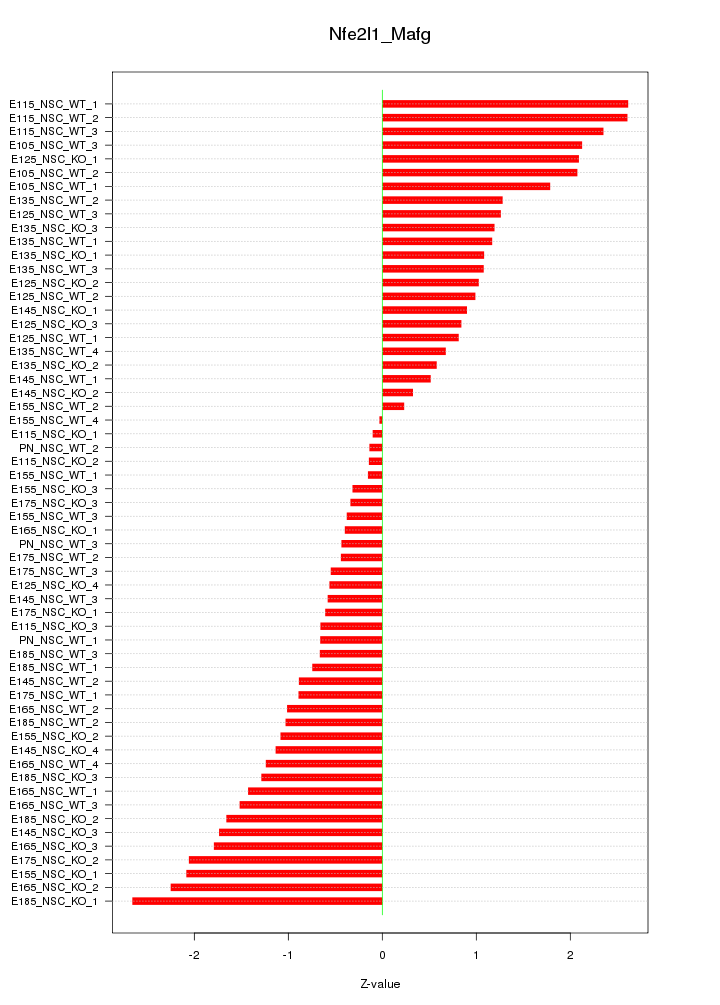
Motif ID: Nfe2l1_Mafg
Z-value: 1.282

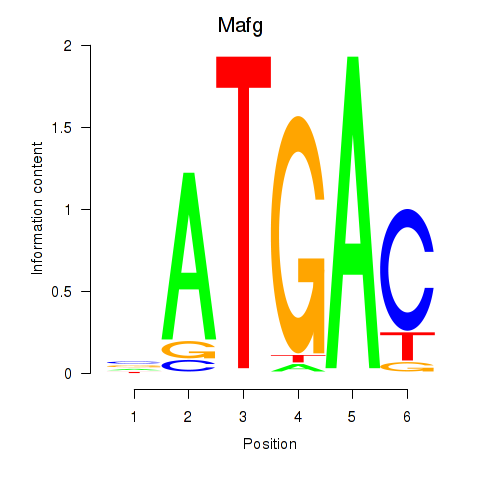
Transcription factors associated with Nfe2l1_Mafg:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Mafg | ENSMUSG00000051510.7 | Mafg |
| Mafg | ENSMUSG00000053906.4 | Mafg |
| Nfe2l1 | ENSMUSG00000038615.11 | Nfe2l1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mafg | mm10_v2_chr11_-_120630516_120630636 | -0.77 | 1.0e-12 | Click! |
| Nfe2l1 | mm10_v2_chr11_-_96824008_96824159 | 0.21 | 1.1e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.2 | 48.9 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 2.9 | 8.7 | GO:0021759 | globus pallidus development(GO:0021759) |
| 1.6 | 4.8 | GO:0035604 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) coronal suture morphogenesis(GO:0060365) squamous basal epithelial stem cell differentiation involved in prostate gland acinus development(GO:0060529) |
| 1.1 | 4.4 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 1.1 | 7.5 | GO:0051461 | regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 1.0 | 3.9 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 1.0 | 4.8 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.9 | 4.7 | GO:0097475 | motor neuron migration(GO:0097475) spinal cord motor neuron migration(GO:0097476) |
| 0.8 | 2.5 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.8 | 6.2 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.8 | 4.6 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.7 | 2.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.6 | 1.8 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.6 | 1.7 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.6 | 1.7 | GO:0071649 | regulation of chemokine (C-C motif) ligand 5 production(GO:0071649) positive regulation of somatostatin secretion(GO:0090274) |
| 0.6 | 1.7 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.5 | 2.5 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.5 | 3.0 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.5 | 2.9 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.5 | 3.7 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.5 | 2.8 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.4 | 2.6 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.4 | 2.1 | GO:0090164 | protein retention in Golgi apparatus(GO:0045053) asymmetric Golgi ribbon formation(GO:0090164) |
| 0.4 | 1.1 | GO:0002842 | T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) positive regulation of T cell mediated immune response to tumor cell(GO:0002842) |
| 0.4 | 1.8 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.3 | 2.0 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.3 | 4.4 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.3 | 3.6 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.3 | 2.2 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.3 | 1.6 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.3 | 1.5 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.3 | 7.0 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.3 | 2.8 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.3 | 4.9 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.3 | 0.8 | GO:2000832 | cortisol secretion(GO:0043400) regulation of cortisol secretion(GO:0051462) negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.3 | 0.8 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.2 | 0.7 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.2 | 1.4 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.2 | 6.8 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.2 | 2.7 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.2 | 1.5 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.2 | 2.2 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.2 | 1.5 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.2 | 0.5 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.2 | 0.7 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.2 | 3.1 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.2 | 0.5 | GO:0030210 | heparin biosynthetic process(GO:0030210) Tie signaling pathway(GO:0048014) |
| 0.2 | 3.4 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.2 | 0.8 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 | 0.4 | GO:0007210 | serotonin receptor signaling pathway(GO:0007210) |
| 0.1 | 1.2 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.1 | 0.8 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 0.6 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 2.5 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 0.7 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.1 | 2.3 | GO:0006625 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.1 | 0.4 | GO:0015786 | UDP-glucose transport(GO:0015786) |
| 0.1 | 2.2 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.1 | 2.3 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 | 0.6 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.1 | 3.3 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.3 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 0.1 | 1.2 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 | 0.3 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.1 | 1.6 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.1 | 0.4 | GO:0072368 | regulation of lipid transport by negative regulation of transcription from RNA polymerase II promoter(GO:0072368) |
| 0.1 | 2.2 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.1 | 6.0 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.1 | 5.8 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.1 | 1.0 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.1 | 0.3 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 0.6 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.1 | 4.3 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 2.9 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.1 | 0.3 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.5 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.9 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 1.0 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 7.5 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.1 | 1.7 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.1 | 1.0 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.1 | 7.1 | GO:0000077 | DNA damage checkpoint(GO:0000077) |
| 0.1 | 0.6 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 1.8 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 2.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 0.6 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.0 | 0.9 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 1.0 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 2.3 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 1.4 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.0 | 1.6 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 1.2 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.7 | GO:0003014 | renal system process(GO:0003014) |
| 0.0 | 0.6 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 5.0 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.8 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.0 | 2.1 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 2.0 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 1.0 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 6.8 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 1.2 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.9 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.8 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 1.0 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 2.1 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 2.1 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.2 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.3 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.5 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 2.3 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 1.9 | GO:0042113 | B cell activation(GO:0042113) |
| 0.0 | 0.6 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 0.5 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 7.5 | GO:0008623 | CHRAC(GO:0008623) |
| 0.9 | 2.7 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.9 | 7.0 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.8 | 3.1 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.6 | 1.8 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.5 | 2.1 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.4 | 2.3 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.4 | 2.6 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.3 | 4.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.3 | 1.7 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.3 | 2.9 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.3 | 0.9 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.3 | 2.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.3 | 1.8 | GO:0033503 | HULC complex(GO:0033503) |
| 0.2 | 3.5 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.2 | 4.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.2 | 2.0 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.2 | 2.0 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.2 | 3.0 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.2 | 1.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) lipopolysaccharide receptor complex(GO:0046696) |
| 0.2 | 2.1 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.9 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.1 | 1.8 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 0.9 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 1.4 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 5.0 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.1 | 0.8 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 1.0 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 4.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 1.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 4.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.4 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 0.4 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 3.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 6.8 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 18.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.2 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 2.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 3.1 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.2 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 2.9 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.8 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.7 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 2.1 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 1.4 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.8 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.9 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.6 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.2 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.3 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.3 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 1.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.5 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.6 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 1.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 1.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.9 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 4.3 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.1 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.6 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.3 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.8 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.8 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.9 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 7.0 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.9 | 4.4 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.9 | 3.4 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.8 | 7.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.8 | 4.9 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.8 | 2.5 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.7 | 3.0 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.7 | 8.7 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.6 | 3.1 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.6 | 7.0 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.5 | 4.8 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.5 | 2.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.5 | 1.5 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.5 | 2.3 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.4 | 3.7 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.4 | 2.8 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.4 | 4.9 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.3 | 1.0 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.3 | 3.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.3 | 6.1 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.3 | 4.2 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.3 | 1.5 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) |
| 0.3 | 2.6 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.3 | 6.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.3 | 1.4 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.3 | 1.4 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.3 | 1.0 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) prostaglandin-E synthase activity(GO:0050220) |
| 0.3 | 53.2 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.2 | 1.7 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.2 | 2.9 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.2 | 0.6 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.2 | 1.4 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.2 | 4.9 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.2 | 4.8 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.2 | 0.5 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.2 | 0.8 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.2 | 0.6 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.2 | 7.3 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.2 | 4.7 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.2 | 2.0 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 1.8 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 1.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 0.8 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 1.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 5.0 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 4.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.8 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 0.4 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin binding(GO:0051378) serotonin receptor activity(GO:0099589) |
| 0.1 | 3.1 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.1 | 1.9 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.4 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 1.7 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.7 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 2.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.6 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.8 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 2.9 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) |
| 0.1 | 3.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 1.0 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 14.5 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.1 | 4.1 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 1.7 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 1.0 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 2.7 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.7 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.0 | 1.0 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 1.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 3.3 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.0 | 0.2 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 2.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.2 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.0 | 0.2 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 1.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.9 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 1.6 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.3 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 2.9 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 1.5 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) |
| 0.0 | 6.3 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 6.6 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.4 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.6 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.7 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 2.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |