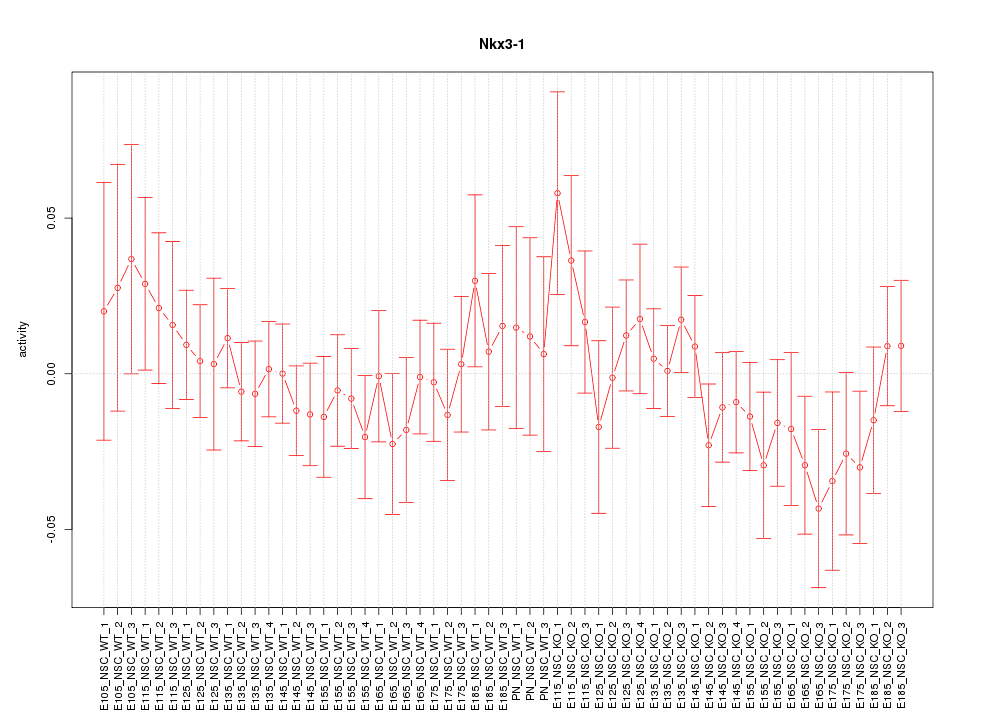
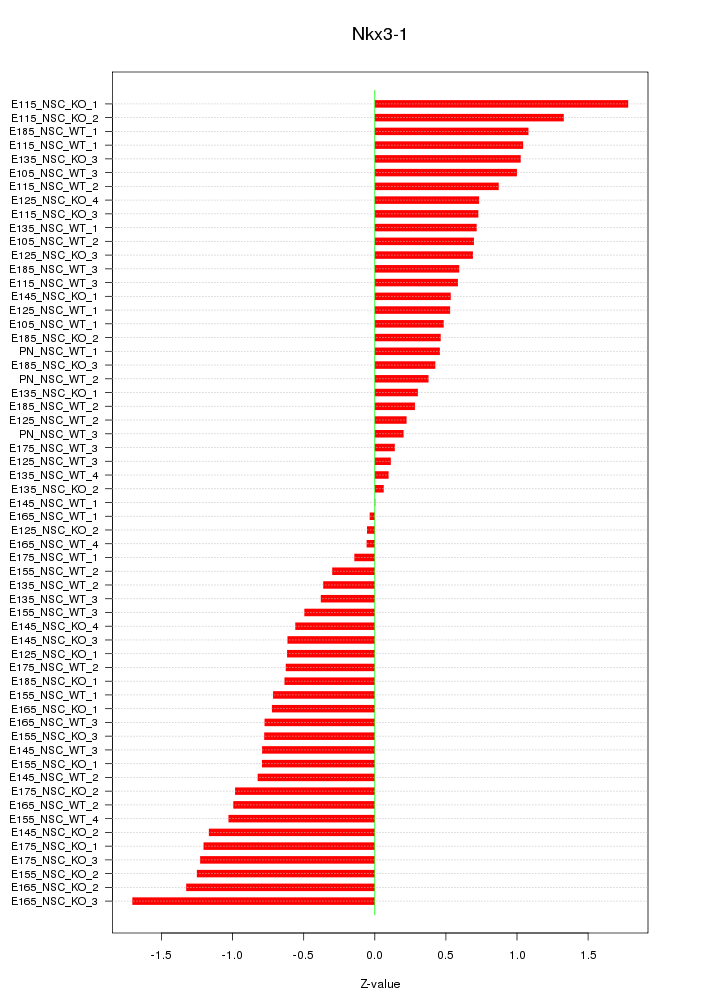
Motif ID: Nkx3-1
Z-value: 0.774
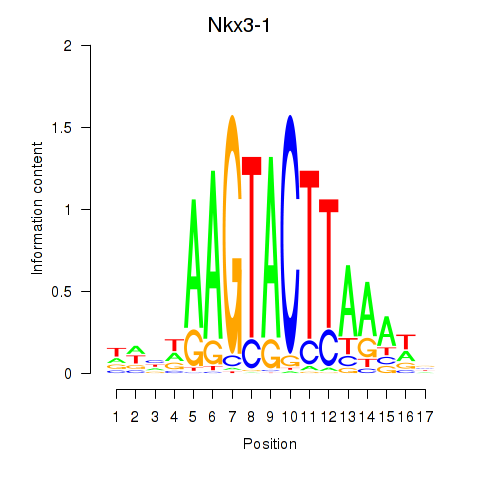
Transcription factors associated with Nkx3-1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nkx3-1 | ENSMUSG00000022061.8 | Nkx3-1 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.5 | GO:0015938 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) |
| 1.5 | 4.4 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 1.3 | 4.0 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.8 | 5.9 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.8 | 7.2 | GO:0019240 | citrulline biosynthetic process(GO:0019240) |
| 0.8 | 5.6 | GO:0000022 | mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.8 | 2.3 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 0.7 | 2.2 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.7 | 2.9 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.7 | 5.0 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.7 | 2.1 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.7 | 2.0 | GO:0002765 | immune response-inhibiting signal transduction(GO:0002765) |
| 0.6 | 3.2 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.6 | 1.8 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) glucose 1-phosphate metabolic process(GO:0019255) |
| 0.5 | 8.4 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.5 | 4.0 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 0.5 | 1.9 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.4 | 3.1 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.4 | 3.3 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.4 | 5.0 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.4 | 1.8 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.3 | 2.7 | GO:1901550 | regulation of endothelial cell development(GO:1901550) regulation of establishment of endothelial barrier(GO:1903140) |
| 0.3 | 2.0 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.3 | 2.6 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.3 | 1.2 | GO:2000407 | positive regulation of necrotic cell death(GO:0010940) regulation of T cell extravasation(GO:2000407) |
| 0.3 | 0.9 | GO:0046101 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.2 | 3.7 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.2 | 1.5 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) amylase secretion(GO:0036394) |
| 0.2 | 6.1 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.2 | 0.9 | GO:0010920 | negative regulation of inositol phosphate biosynthetic process(GO:0010920) |
| 0.2 | 1.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.2 | 3.0 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.2 | 0.9 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.2 | 0.7 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.2 | 3.9 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.2 | 1.8 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 1.8 | GO:0070633 | transepithelial transport(GO:0070633) |
| 0.1 | 1.9 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.1 | 1.2 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.1 | 1.1 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.1 | 0.4 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.1 | 0.7 | GO:1900116 | sequestering of extracellular ligand from receptor(GO:0035581) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 | 1.4 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 | 2.4 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.1 | 0.3 | GO:2000642 | negative regulation of vacuolar transport(GO:1903336) negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.1 | 0.9 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 0.2 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 0.6 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.1 | 2.6 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.1 | 1.2 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.1 | 1.2 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 1.9 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.1 | 0.8 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) ceramide catabolic process(GO:0046514) sphingoid biosynthetic process(GO:0046520) |
| 0.1 | 2.0 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 0.5 | GO:0009301 | snRNA transcription(GO:0009301) |
| 0.1 | 2.8 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.1 | 0.6 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 1.6 | GO:0071158 | positive regulation of cell cycle arrest(GO:0071158) |
| 0.1 | 1.6 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.1 | 1.6 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 0.2 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.0 | 1.6 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 2.2 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 1.2 | GO:2000279 | negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.0 | 0.3 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 1.1 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 6.2 | GO:0060348 | bone development(GO:0060348) |
| 0.0 | 0.9 | GO:0090382 | phagosome maturation(GO:0090382) |
| 0.0 | 0.4 | GO:0006750 | glutathione biosynthetic process(GO:0006750) response to cadmium ion(GO:0046686) |
| 0.0 | 0.4 | GO:0010388 | protein deneddylation(GO:0000338) cullin deneddylation(GO:0010388) |
| 0.0 | 0.4 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 1.9 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 0.2 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.0 | 1.6 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.6 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.4 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 0.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.7 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 0.8 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.1 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 1.0 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.3 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.6 | 1.9 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.6 | 1.8 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.5 | 1.9 | GO:0005712 | chiasma(GO:0005712) |
| 0.3 | 2.6 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.3 | 4.0 | GO:0070938 | contractile ring(GO:0070938) |
| 0.3 | 0.9 | GO:0000811 | GINS complex(GO:0000811) |
| 0.2 | 3.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.2 | 0.9 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.2 | 1.4 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.2 | 7.8 | GO:0032420 | stereocilium(GO:0032420) |
| 0.2 | 1.2 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.1 | 1.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 8.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 1.6 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 2.0 | GO:0042611 | MHC protein complex(GO:0042611) |
| 0.1 | 3.0 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 0.7 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 1.1 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.1 | 5.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.6 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.1 | 1.0 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.4 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 3.7 | GO:0000791 | euchromatin(GO:0000791) |
| 0.1 | 3.9 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 6.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 5.0 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.0 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.4 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 9.0 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.3 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 1.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 2.0 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.5 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 1.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.5 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 1.2 | 3.7 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 1.2 | 7.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.9 | 7.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.8 | 2.3 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.6 | 5.0 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.5 | 6.3 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.5 | 1.9 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.5 | 1.9 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.5 | 2.9 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.3 | 5.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.3 | 1.8 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.3 | 6.8 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.3 | 0.8 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.2 | 2.6 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 4.0 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.2 | 0.9 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.2 | 1.6 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.2 | 1.8 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.2 | 2.0 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.2 | 1.6 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 1.8 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 3.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.2 | 1.6 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.2 | 5.0 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 2.2 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 3.0 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 2.0 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 3.2 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 0.3 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.7 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 2.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 0.7 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.1 | 2.1 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 1.5 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 1.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 1.2 | GO:0016594 | glycine binding(GO:0016594) |
| 0.1 | 1.3 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.1 | 1.2 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.1 | 2.8 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 2.0 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 1.1 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.1 | 0.5 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.9 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 1.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 2.6 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 7.4 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.2 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.0 | 0.9 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.6 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.0 | 1.2 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.6 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.0 | 0.8 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 1.7 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.1 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) |
| 0.0 | 0.4 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 2.7 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.5 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 3.8 | GO:0000982 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 2.9 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |