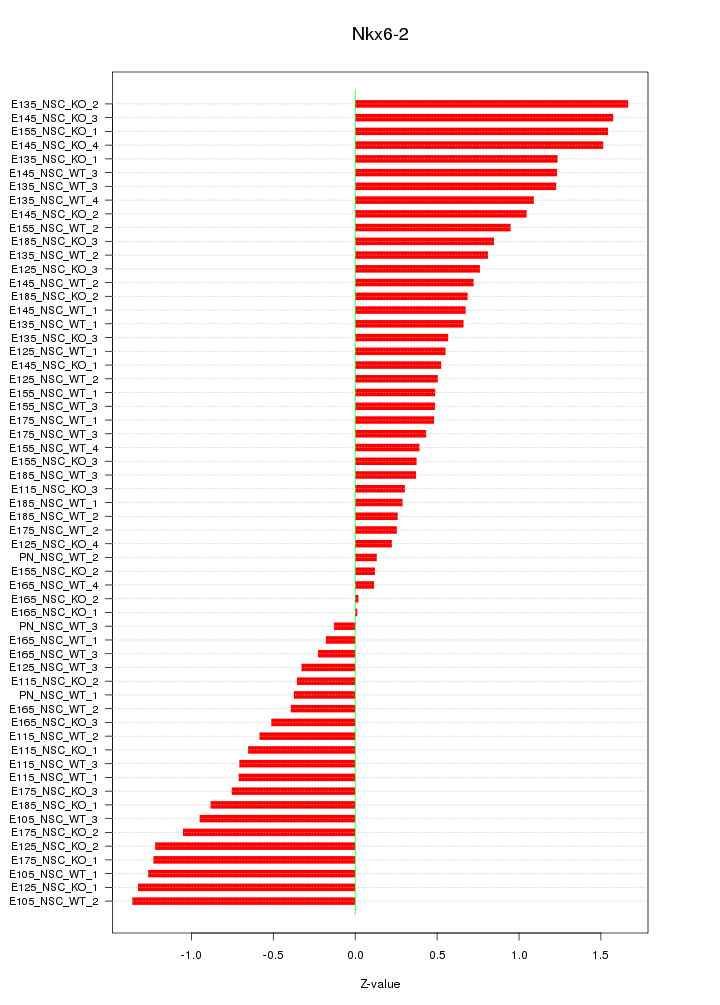
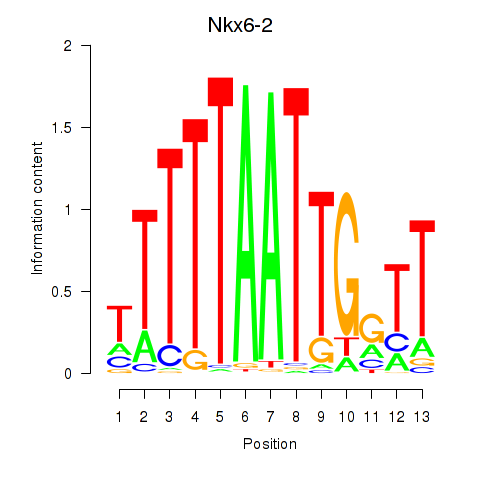
Motif ID: Nkx6-2
Z-value: 0.809
Transcription factors associated with Nkx6-2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nkx6-2 | ENSMUSG00000041309.11 | Nkx6-2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx6-2 | mm10_v2_chr7_-_139582790_139582808 | -0.43 | 5.9e-04 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 8.4 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 1.0 | 3.0 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.8 | 5.4 | GO:0034047 | regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.7 | 1.4 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.7 | 3.9 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.6 | 1.9 | GO:0038095 | positive regulation of mast cell cytokine production(GO:0032765) Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.6 | 5.9 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.5 | 2.0 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.4 | 4.7 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.4 | 1.7 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) negative regulation of respiratory burst(GO:0060268) |
| 0.4 | 1.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.3 | 2.2 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.3 | 2.4 | GO:2000809 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.3 | 1.8 | GO:1903056 | regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.3 | 1.3 | GO:0014719 | skeletal muscle satellite cell activation(GO:0014719) |
| 0.3 | 1.0 | GO:0050913 | sensory perception of bitter taste(GO:0050913) |
| 0.2 | 2.8 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 0.6 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 0.7 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 1.5 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.1 | 1.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.4 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.1 | 0.8 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 1.3 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.1 | 7.7 | GO:0021954 | central nervous system neuron development(GO:0021954) |
| 0.1 | 0.5 | GO:0006344 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) maintenance of chromatin silencing(GO:0006344) |
| 0.1 | 0.6 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.4 | GO:1902965 | protein localization to early endosome(GO:1902946) regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.1 | 0.3 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.6 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 1.0 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 1.6 | GO:0051926 | negative regulation of calcium ion transport(GO:0051926) |
| 0.0 | 0.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.6 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.5 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.2 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.2 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 | 1.5 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.0 | 0.6 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 1.9 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 1.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.3 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.0 | 1.2 | GO:0006096 | glycolytic process(GO:0006096) ATP generation from ADP(GO:0006757) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 5.9 | GO:0005883 | neurofilament(GO:0005883) |
| 0.6 | 5.4 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.5 | 2.4 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.4 | 4.7 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.2 | 4.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 1.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.6 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 3.0 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 3.9 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 1.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 0.7 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 2.0 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 1.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 8.4 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.1 | 0.9 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 1.4 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 1.5 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.4 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 2.8 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 1.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 2.8 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 2.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 2.0 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 3.2 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 5.4 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.5 | 1.5 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.4 | 8.1 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.4 | 2.4 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.4 | 5.4 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.3 | 4.7 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.3 | 0.9 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.3 | 2.0 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.3 | 1.6 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.2 | 4.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.4 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.1 | 0.6 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.1 | 1.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 1.9 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 1.0 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 1.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 3.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 2.8 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.1 | 1.1 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.1 | 1.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 1.8 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 1.7 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 2.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.3 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.0 | 5.2 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.7 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.9 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.9 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 9.7 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.5 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.4 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 0.6 | GO:0019003 | GDP binding(GO:0019003) |