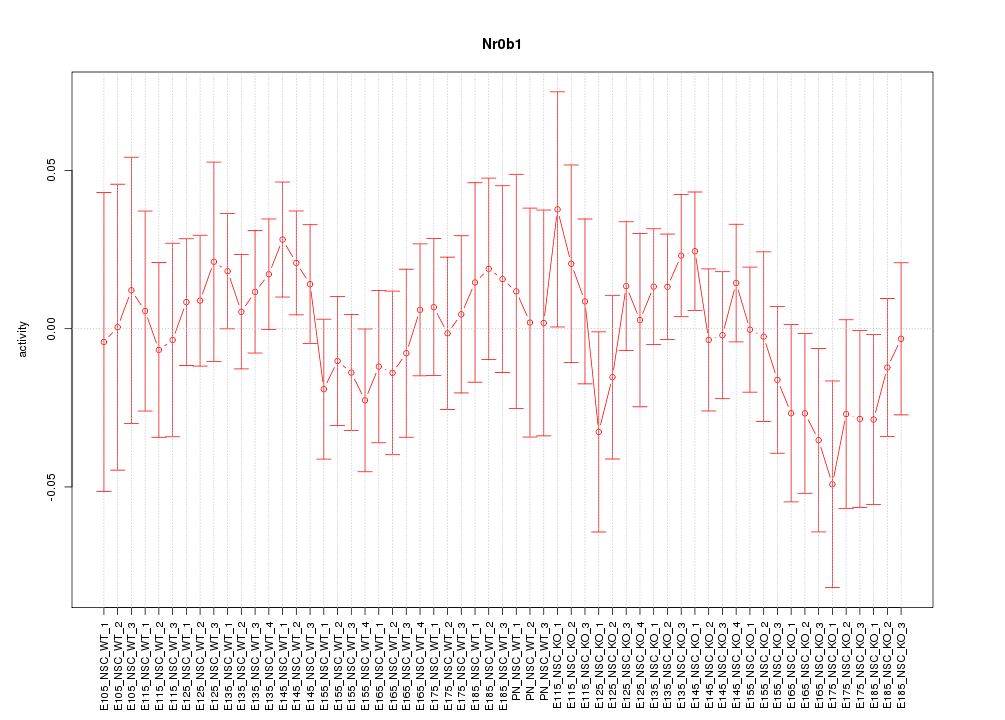
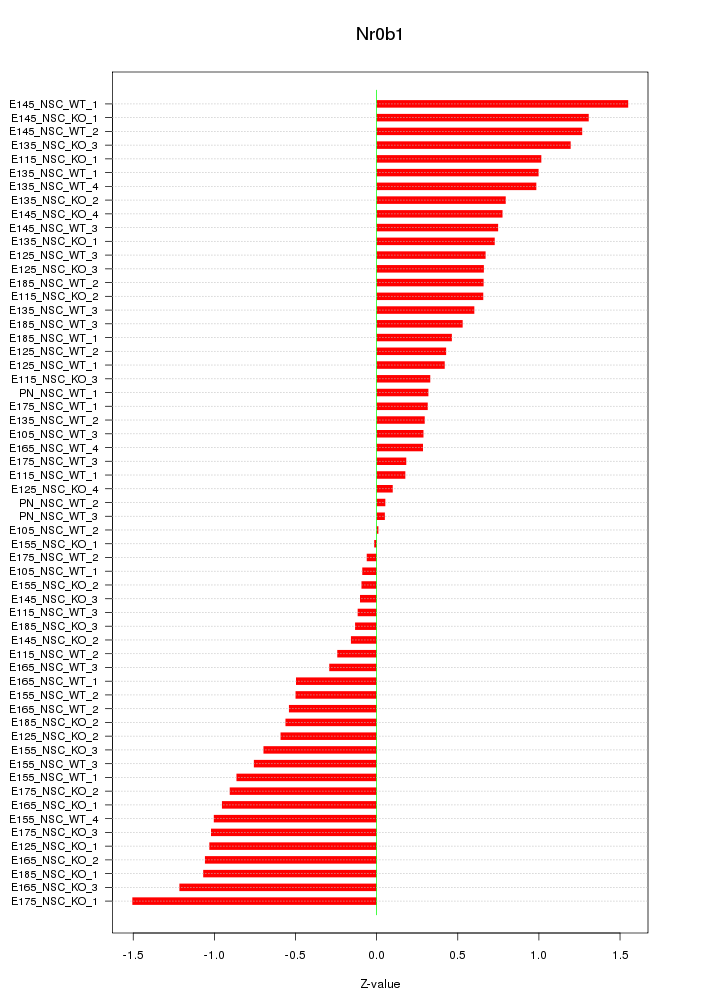
Motif ID: Nr0b1
Z-value: 0.718
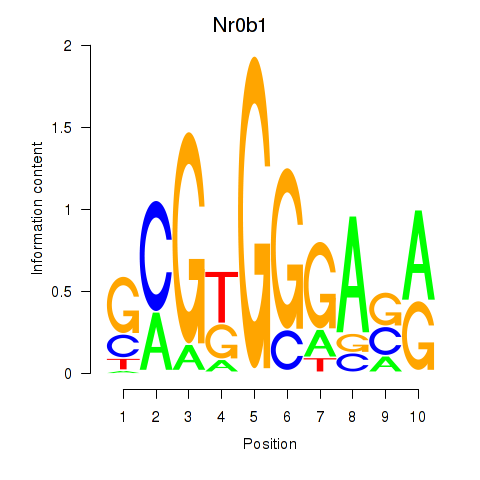
Transcription factors associated with Nr0b1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nr0b1 | ENSMUSG00000025056.4 | Nr0b1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr0b1 | mm10_v2_chrX_+_86191764_86191782 | -0.28 | 2.9e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 8.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.7 | 8.0 | GO:0036289 | peptidyl-serine autophosphorylation(GO:0036289) |
| 0.5 | 2.6 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.5 | 6.2 | GO:0014842 | regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.5 | 3.5 | GO:0051461 | positive regulation of heat generation(GO:0031652) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.4 | 1.3 | GO:0072070 | loop of Henle development(GO:0072070) metanephric loop of Henle development(GO:0072236) |
| 0.4 | 2.0 | GO:0007223 | muscular septum morphogenesis(GO:0003150) Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.4 | 1.5 | GO:0035878 | nail development(GO:0035878) |
| 0.4 | 1.5 | GO:2001045 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.4 | 1.4 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.3 | 1.7 | GO:0051661 | maintenance of centrosome location(GO:0051661) regulation of microtubule motor activity(GO:2000574) |
| 0.3 | 2.5 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.3 | 1.1 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.2 | 0.7 | GO:1902915 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.2 | 3.8 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.2 | 0.9 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.2 | 0.5 | GO:0007354 | zygotic determination of anterior/posterior axis, embryo(GO:0007354) |
| 0.2 | 1.1 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.2 | 0.3 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 1.0 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.1 | 0.6 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.1 | 1.4 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 1.1 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 0.5 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 | 1.3 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 0.3 | GO:0036166 | phenotypic switching(GO:0036166) |
| 0.1 | 0.3 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 1.0 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 0.4 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.1 | 1.1 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 0.8 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.1 | 0.5 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.1 | 0.2 | GO:0045759 | negative regulation of action potential(GO:0045759) |
| 0.0 | 2.4 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.5 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.4 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.9 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.2 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 2.3 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 3.1 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.6 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.2 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 0.8 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.3 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.2 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 8.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.3 | 1.7 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.3 | 1.1 | GO:0002111 | BRCA2-BRAF35 complex(GO:0002111) |
| 0.2 | 1.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 1.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 1.5 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.9 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 0.6 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 10.9 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 1.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 1.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.8 | GO:0005581 | collagen trimer(GO:0005581) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 8.0 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.5 | 1.5 | GO:0071862 | protein phosphatase type 1 activator activity(GO:0071862) |
| 0.4 | 3.0 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.4 | 1.9 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.3 | 0.9 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.3 | 1.7 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.2 | 1.6 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.2 | 3.5 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.2 | 3.8 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 1.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.3 | GO:0048045 | 4-hydroxybenzoate octaprenyltransferase activity(GO:0008412) protoheme IX farnesyltransferase activity(GO:0008495) (S)-2,3-di-O-geranylgeranylglyceryl phosphate synthase activity(GO:0043888) cadaverine aminopropyltransferase activity(GO:0043918) agmatine aminopropyltransferase activity(GO:0043919) 1,4-dihydroxy-2-naphthoate octaprenyltransferase activity(GO:0046428) trans-pentaprenyltranstransferase activity(GO:0048045) ATP dimethylallyltransferase activity(GO:0052622) ADP dimethylallyltransferase activity(GO:0052623) |
| 0.1 | 2.0 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 2.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.2 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 1.1 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 0.7 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.3 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 1.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 1.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 1.0 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.5 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 2.2 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.9 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.2 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 4.5 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 1.2 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 2.5 | GO:0001047 | core promoter binding(GO:0001047) |