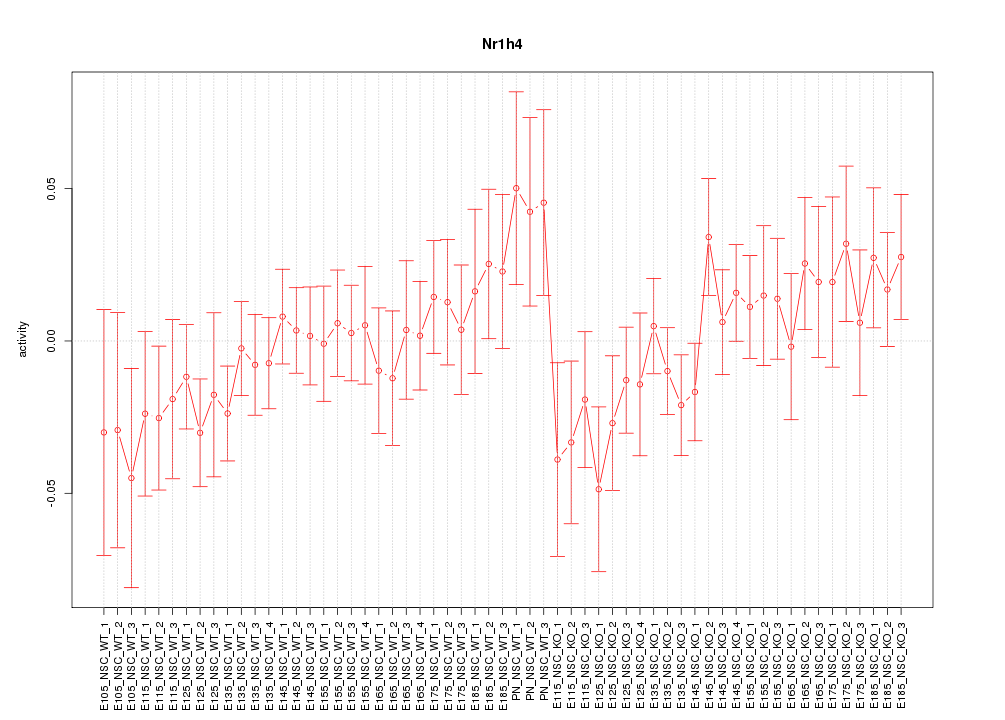
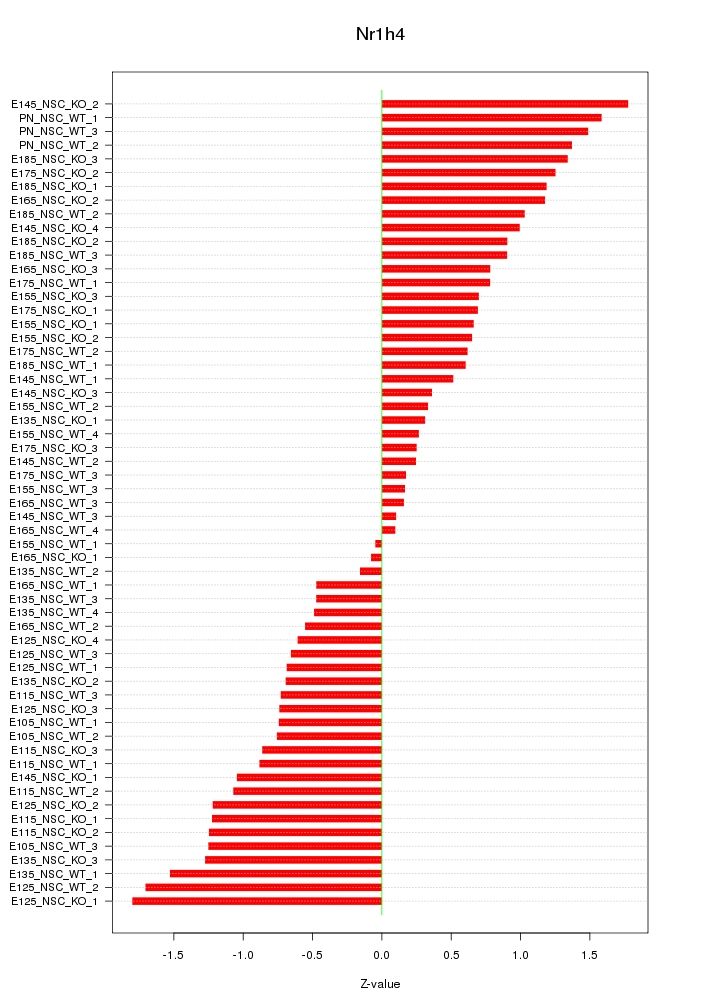
Motif ID: Nr1h4
Z-value: 0.914
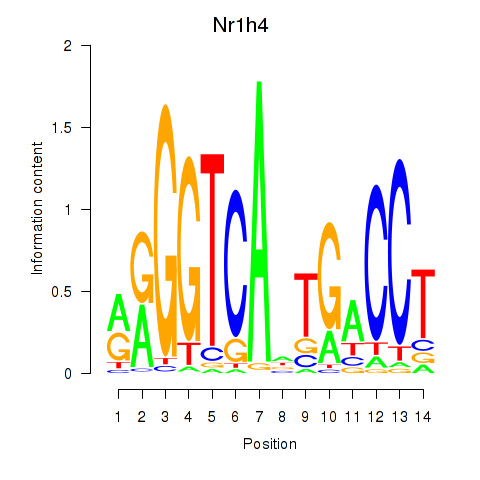
Transcription factors associated with Nr1h4:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nr1h4 | ENSMUSG00000047638.9 | Nr1h4 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.8 | GO:1903795 | regulation of inorganic anion transmembrane transport(GO:1903795) |
| 1.9 | 5.7 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 1.9 | 5.7 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 1.7 | 5.1 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 1.7 | 5.1 | GO:0043465 | regulation of fermentation(GO:0043465) negative regulation of fermentation(GO:1901003) |
| 1.6 | 4.9 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 1.6 | 6.3 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 1.5 | 8.8 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 1.2 | 8.6 | GO:0010273 | detoxification of copper ion(GO:0010273) cellular response to zinc ion(GO:0071294) stress response to copper ion(GO:1990169) |
| 1.1 | 3.4 | GO:1904049 | regulation of spontaneous neurotransmitter secretion(GO:1904048) negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 1.0 | 3.9 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.8 | 2.4 | GO:1903525 | regulation of membrane tubulation(GO:1903525) |
| 0.8 | 3.8 | GO:0015871 | choline transport(GO:0015871) |
| 0.7 | 3.0 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.7 | 2.1 | GO:1902071 | positive regulation of cellular response to hypoxia(GO:1900039) regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.6 | 1.9 | GO:0019401 | alditol biosynthetic process(GO:0019401) |
| 0.6 | 1.8 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.5 | 2.0 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.4 | 3.8 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.4 | 2.3 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.4 | 1.8 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.3 | 3.7 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.3 | 8.9 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.3 | 0.9 | GO:1903903 | regulation of establishment of T cell polarity(GO:1903903) |
| 0.3 | 2.9 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.3 | 13.5 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.3 | 1.6 | GO:0046880 | regulation of follicle-stimulating hormone secretion(GO:0046880) follicle-stimulating hormone secretion(GO:0046884) |
| 0.2 | 1.2 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.2 | 1.4 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.2 | 0.9 | GO:0060745 | mammary gland branching involved in pregnancy(GO:0060745) |
| 0.2 | 1.0 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.2 | 0.6 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.2 | 1.5 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 4.2 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.1 | 0.5 | GO:1903546 | microtubule anchoring at centrosome(GO:0034454) negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 | 7.6 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.1 | 0.4 | GO:0045414 | regulation of interleukin-8 biosynthetic process(GO:0045414) ERK5 cascade(GO:0070375) |
| 0.1 | 0.5 | GO:0006742 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.1 | 2.6 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.1 | 0.1 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.1 | 2.1 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 | 2.5 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.0 | 7.4 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 2.0 | GO:0010771 | negative regulation of cell morphogenesis involved in differentiation(GO:0010771) |
| 0.0 | 0.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 3.1 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 3.3 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.4 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 5.8 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.0 | 2.7 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 1.6 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.4 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.0 | 2.7 | GO:0006096 | glycolytic process(GO:0006096) ATP generation from ADP(GO:0006757) |
| 0.0 | 0.9 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.0 | 1.0 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.4 | GO:0051974 | negative regulation of telomerase activity(GO:0051974) |
| 0.0 | 0.3 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 0.6 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.3 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 1.0 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 2.1 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 1.7 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.6 | GO:0021795 | cerebral cortex cell migration(GO:0021795) |
| 0.0 | 0.9 | GO:0006073 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.8 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.7 | 4.2 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.7 | 3.4 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.5 | 2.7 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.5 | 1.6 | GO:0043512 | inhibin-betaglycan-ActRII complex(GO:0034673) inhibin A complex(GO:0043512) |
| 0.5 | 5.7 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.3 | 1.8 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.3 | 0.9 | GO:1990879 | CST complex(GO:1990879) |
| 0.3 | 1.8 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.3 | 2.3 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.2 | 7.6 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.2 | 8.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.2 | 1.5 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.1 | 3.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 2.0 | GO:0005773 | vacuole(GO:0005773) |
| 0.1 | 0.9 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 1.2 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.9 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 7.9 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 0.7 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 0.8 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.1 | 0.3 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.1 | 6.3 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 1.0 | GO:0030123 | AP-3 adaptor complex(GO:0030123) HOPS complex(GO:0030897) |
| 0.1 | 2.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.8 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.9 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.5 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 5.1 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 1.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 2.7 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.7 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 2.4 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 3.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 7.4 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 5.3 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 1.4 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 5.1 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 5.6 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 3.0 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 1.4 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.6 | GO:0031526 | brush border membrane(GO:0031526) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.7 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 1.4 | 5.7 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 1.1 | 6.3 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 1.0 | 8.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 1.0 | 3.8 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.8 | 5.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.8 | 4.9 | GO:0009374 | biotin binding(GO:0009374) |
| 0.8 | 3.8 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.7 | 3.0 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.7 | 3.7 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.7 | 2.6 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.5 | 1.9 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.4 | 2.9 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.4 | 2.8 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.3 | 1.4 | GO:0004096 | catalase activity(GO:0004096) |
| 0.3 | 0.9 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.3 | 2.7 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.3 | 0.9 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.2 | 2.7 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.2 | 3.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 1.5 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.2 | 2.3 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.2 | 8.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 0.9 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.2 | 9.4 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.2 | 1.6 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.2 | 8.6 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 0.6 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.1 | 0.9 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 3.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 2.5 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 7.4 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 6.4 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.1 | 3.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.8 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 2.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 0.9 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 0.5 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.6 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.1 | 2.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 5.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 0.4 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 3.9 | GO:0061733 | peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.0 | 2.4 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.5 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.0 | 2.0 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 1.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.4 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.4 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 3.9 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 3.1 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 1.6 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.9 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 1.0 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 2.4 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.3 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.0 | 0.8 | GO:0035496 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) O antigen polymerase activity(GO:0008755) lipopolysaccharide-1,6-galactosyltransferase activity(GO:0008921) cellulose synthase activity(GO:0016759) 9-phenanthrol UDP-glucuronosyltransferase activity(GO:0018715) 1-phenanthrol glycosyltransferase activity(GO:0018716) 9-phenanthrol glycosyltransferase activity(GO:0018717) 1,2-dihydroxy-phenanthrene glycosyltransferase activity(GO:0018718) phenanthrol glycosyltransferase activity(GO:0019112) alpha-1,2-galactosyltransferase activity(GO:0031278) dolichyl pyrophosphate Man7GlcNAc2 alpha-1,3-glucosyltransferase activity(GO:0033556) endogalactosaminidase activity(GO:0033931) lipopolysaccharide-1,5-galactosyltransferase activity(GO:0035496) dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase activity(GO:0042283) inositol phosphoceramide synthase activity(GO:0045140) alpha-(1->3)-fucosyltransferase activity(GO:0046920) indole-3-butyrate beta-glucosyltransferase activity(GO:0052638) salicylic acid glucosyltransferase (ester-forming) activity(GO:0052639) salicylic acid glucosyltransferase (glucoside-forming) activity(GO:0052640) benzoic acid glucosyltransferase activity(GO:0052641) chondroitin hydrolase activity(GO:0052757) dolichyl-pyrophosphate Man7GlcNAc2 alpha-1,6-mannosyltransferase activity(GO:0052824) cytokinin 9-beta-glucosyltransferase activity(GO:0080062) |
| 0.0 | 0.1 | GO:0043842 | Kdo transferase activity(GO:0043842) |