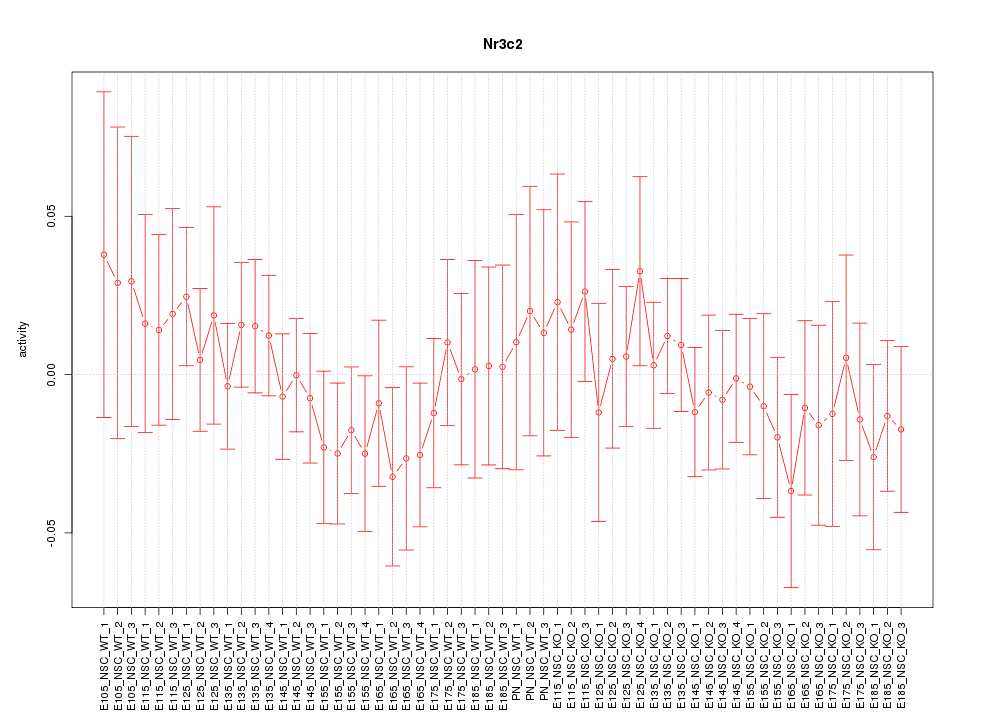
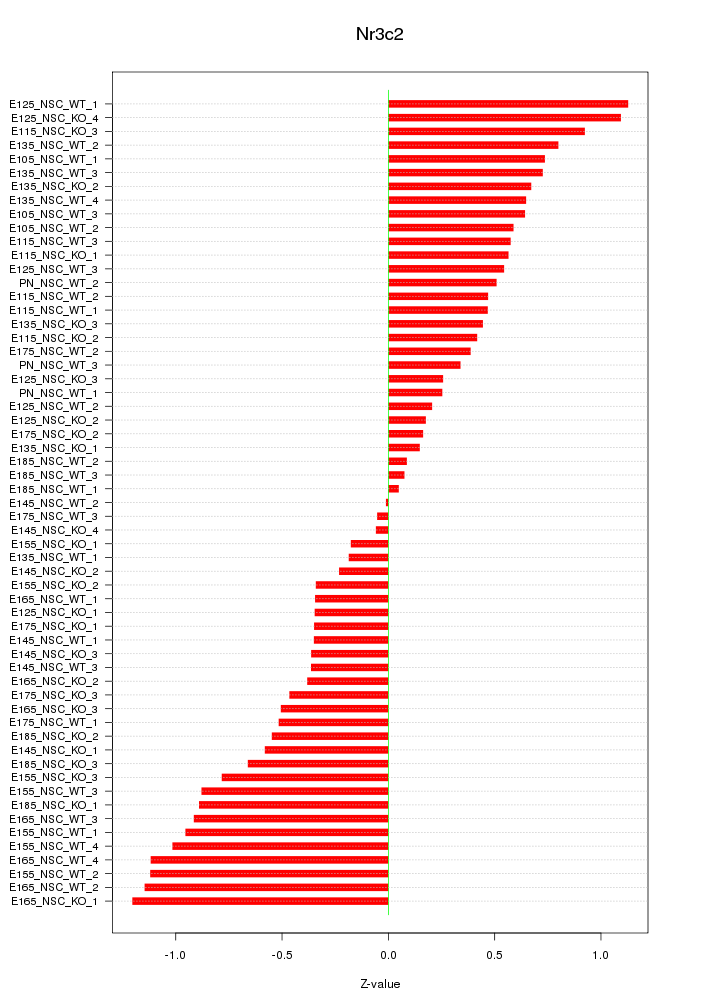
Motif ID: Nr3c2
Z-value: 0.616

Transcription factors associated with Nr3c2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nr3c2 | ENSMUSG00000031618.7 | Nr3c2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr3c2 | mm10_v2_chr8_+_76902277_76902476 | -0.44 | 4.3e-04 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.2 | GO:0042822 | vitamin B6 metabolic process(GO:0042816) pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.9 | 2.6 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.8 | 6.4 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.6 | 5.0 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.5 | 1.4 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.4 | 3.0 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.4 | 2.6 | GO:0032782 | cellular water homeostasis(GO:0009992) bile acid secretion(GO:0032782) |
| 0.2 | 1.5 | GO:0010273 | detoxification of copper ion(GO:0010273) cellular response to zinc ion(GO:0071294) stress response to copper ion(GO:1990169) |
| 0.1 | 3.9 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 5.1 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 1.4 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 2.5 | GO:0017157 | regulation of exocytosis(GO:0017157) |
| 0.0 | 0.2 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.4 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.0 | 2.2 | GO:0008360 | regulation of cell shape(GO:0008360) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.0 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.5 | 2.6 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 2.8 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 0.4 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.2 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 2.0 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.0 | 2.5 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 1.5 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.2 | GO:0031403 | lithium ion binding(GO:0031403) |
| 0.5 | 6.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.3 | 2.6 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) glycerol channel activity(GO:0015254) |
| 0.2 | 5.1 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.2 | 2.0 | GO:0048038 | quinone binding(GO:0048038) |
| 0.1 | 3.0 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 1.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 3.9 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 2.6 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 0.1 | GO:0034511 | U4 snRNA binding(GO:0030621) U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.9 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |