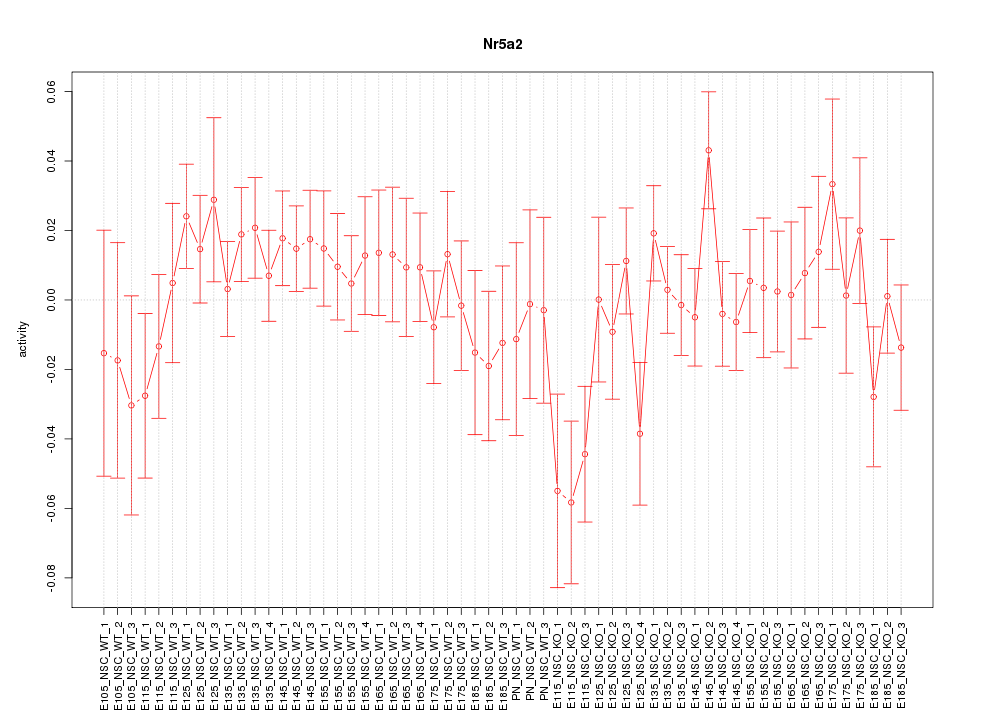
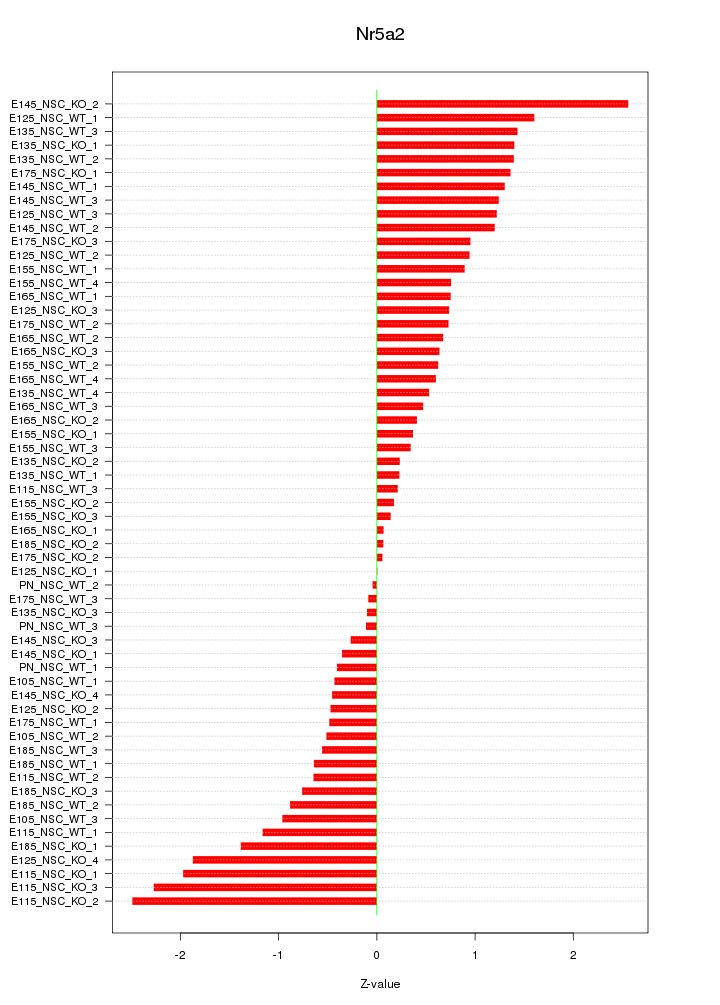
Motif ID: Nr5a2
Z-value: 0.988
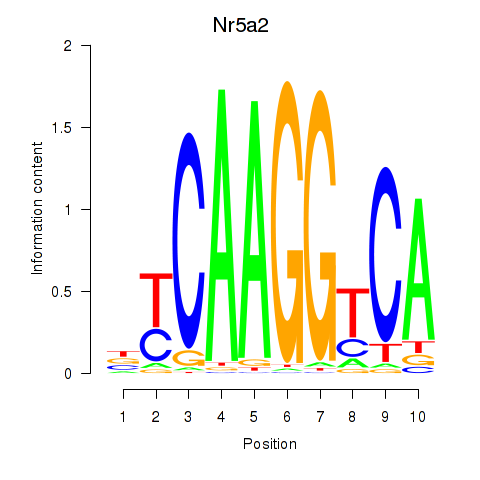
Transcription factors associated with Nr5a2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nr5a2 | ENSMUSG00000026398.8 | Nr5a2 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 14.0 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 2.8 | 8.3 | GO:0021933 | radial glia guided migration of cerebellar granule cell(GO:0021933) |
| 1.7 | 5.2 | GO:1900736 | regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900736) |
| 1.6 | 4.9 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 1.1 | 3.2 | GO:0071544 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 1.1 | 4.2 | GO:0021564 | glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 1.0 | 3.1 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleoside transport(GO:0032242) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 0.9 | 4.6 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.9 | 2.6 | GO:0002447 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.9 | 1.7 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.8 | 3.3 | GO:1901524 | regulation of macromitophagy(GO:1901524) negative regulation of macromitophagy(GO:1901525) |
| 0.7 | 2.2 | GO:0043465 | regulation of fermentation(GO:0043465) negative regulation of fermentation(GO:1901003) |
| 0.7 | 4.2 | GO:0046103 | inosine biosynthetic process(GO:0046103) |
| 0.7 | 3.4 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.6 | 1.9 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.6 | 2.5 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.5 | 2.2 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.5 | 13.8 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.5 | 3.0 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.5 | 1.5 | GO:0042560 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.4 | 1.3 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.4 | 3.5 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.4 | 2.3 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.4 | 40.0 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.4 | 3.3 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 0.3 | 1.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.3 | 1.0 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.3 | 1.2 | GO:2000195 | negative regulation of female gonad development(GO:2000195) |
| 0.3 | 2.1 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.3 | 2.4 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.3 | 0.9 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.3 | 1.4 | GO:0030242 | pexophagy(GO:0030242) |
| 0.3 | 2.5 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.3 | 1.3 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.3 | 7.3 | GO:0010043 | response to zinc ion(GO:0010043) |
| 0.2 | 0.5 | GO:0097252 | B cell chemotaxis(GO:0035754) oligodendrocyte apoptotic process(GO:0097252) |
| 0.2 | 2.0 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.2 | 1.2 | GO:0070305 | response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) |
| 0.2 | 0.6 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.2 | 1.4 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.2 | 0.6 | GO:0071649 | regulation of chemokine (C-C motif) ligand 5 production(GO:0071649) |
| 0.2 | 1.8 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.2 | 1.2 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.2 | 2.6 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.2 | 4.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.2 | 3.0 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.2 | 1.5 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.2 | 0.7 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.2 | 2.1 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.2 | 3.7 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.2 | 0.6 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.2 | 3.5 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.2 | 3.9 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 0.8 | GO:0072539 | T-helper 17 cell differentiation(GO:0072539) |
| 0.1 | 0.4 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.1 | 0.7 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 | 1.1 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.6 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.1 | 2.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 12.5 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 0.3 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.1 | 0.7 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.4 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.1 | 0.4 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 1.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 1.1 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.1 | 1.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.5 | GO:0061718 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glycolytic process through glucose-1-phosphate(GO:0061622) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 1.0 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.1 | 6.6 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.1 | 2.0 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 2.3 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 | 8.1 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 2.3 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 0.2 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.1 | 1.1 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.1 | 1.7 | GO:0030262 | cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.1 | 0.4 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 5.5 | GO:0032091 | negative regulation of protein binding(GO:0032091) |
| 0.1 | 1.2 | GO:0001964 | startle response(GO:0001964) |
| 0.1 | 0.7 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.1 | 0.1 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.1 | 1.0 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.3 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.1 | 1.1 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.7 | GO:0010388 | cullin deneddylation(GO:0010388) |
| 0.0 | 0.4 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 2.5 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 1.2 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.8 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.2 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.0 | 0.1 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.0 | 0.2 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.0 | 1.4 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 5.9 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.0 | 3.2 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.5 | GO:0061162 | establishment of monopolar cell polarity(GO:0061162) |
| 0.0 | 0.2 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 5.8 | GO:0043413 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 0.6 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.4 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.7 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.8 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.9 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.0 | 0.7 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 | 0.7 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.5 | GO:0015991 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 0.2 | GO:0051683 | establishment of Golgi localization(GO:0051683) |
| 0.0 | 0.1 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 0.0 | 1.7 | GO:0098792 | xenophagy(GO:0098792) |
| 0.0 | 0.3 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.3 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.0 | GO:0030167 | proteoglycan catabolic process(GO:0030167) |
| 0.0 | 1.1 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 2.6 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.7 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 2.1 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 1.5 | GO:0030010 | establishment of cell polarity(GO:0030010) |
| 0.0 | 0.2 | GO:0060008 | Sertoli cell differentiation(GO:0060008) |
| 0.0 | 1.0 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.2 | GO:0000394 | RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.0 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.0 | 0.6 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 1.5 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.2 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.7 | 14.0 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.7 | 2.6 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.5 | 4.2 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.5 | 1.6 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.5 | 5.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.5 | 3.5 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.4 | 3.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.4 | 4.7 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.4 | 4.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.4 | 12.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.3 | 1.9 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.3 | 12.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.3 | 0.5 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.2 | 2.2 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.2 | 4.4 | GO:0045259 | proton-transporting ATP synthase complex(GO:0045259) |
| 0.2 | 0.9 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.2 | 1.5 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.2 | 6.6 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.2 | 7.8 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 0.5 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.6 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 2.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 2.1 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 1.9 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.1 | 0.7 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 1.4 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 3.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.4 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 5.3 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 1.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 4.9 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 4.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 2.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 25.1 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.1 | 2.9 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 6.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 0.5 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 0.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 0.5 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 1.4 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 1.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 5.1 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 1.2 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 2.3 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 1.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 5.1 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 1.1 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 1.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.6 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 1.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 2.6 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.7 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.4 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 1.5 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 2.6 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.9 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 2.0 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.6 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 1.6 | 4.9 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 1.4 | 5.5 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 1.0 | 2.9 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 1.0 | 5.8 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.9 | 1.8 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.8 | 13.9 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.7 | 2.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.5 | 3.2 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.5 | 1.5 | GO:0034437 | glycoprotein transporter activity(GO:0034437) |
| 0.5 | 3.5 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.5 | 1.5 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.5 | 2.0 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.4 | 3.1 | GO:0032795 | G-protein coupled adenosine receptor activity(GO:0001609) heterotrimeric G-protein binding(GO:0032795) |
| 0.4 | 3.0 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.4 | 7.7 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.4 | 1.4 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.4 | 12.5 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.3 | 1.7 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.3 | 3.7 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.3 | 2.3 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.3 | 4.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.3 | 4.6 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.3 | 1.1 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.3 | 5.8 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 1.2 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.2 | 0.8 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.2 | 2.5 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.2 | 4.3 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.2 | 1.4 | GO:0015288 | porin activity(GO:0015288) |
| 0.2 | 0.7 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.2 | 1.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.2 | 1.5 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 0.6 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 2.9 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.5 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 1.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 1.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.3 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.1 | 1.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 3.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 1.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 2.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 1.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 9.3 | GO:0005267 | potassium channel activity(GO:0005267) |
| 0.1 | 0.4 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 1.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 1.4 | GO:0042165 | neurotransmitter binding(GO:0042165) |
| 0.1 | 1.1 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) acylglycerol lipase activity(GO:0047372) |
| 0.1 | 4.5 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 0.7 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 2.8 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.1 | 1.6 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 0.6 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 3.0 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.1 | 0.6 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.8 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.7 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 0.5 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 2.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 5.1 | GO:0043851 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) rRNA (uridine-2'-O-)-methyltransferase activity(GO:0008650) rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) selenocysteine methyltransferase activity(GO:0016205) rRNA (adenine) methyltransferase activity(GO:0016433) rRNA (cytosine) methyltransferase activity(GO:0016434) rRNA (guanine) methyltransferase activity(GO:0016435) 1-phenanthrol methyltransferase activity(GO:0018707) protein-arginine N5-methyltransferase activity(GO:0019702) dimethylarsinite methyltransferase activity(GO:0034541) 4,5-dihydroxybenzo(a)pyrene methyltransferase activity(GO:0034807) 1-hydroxypyrene methyltransferase activity(GO:0034931) 1-hydroxy-6-methoxypyrene methyltransferase activity(GO:0034933) demethylmenaquinone methyltransferase activity(GO:0043770) cobalt-precorrin-6B C5-methyltransferase activity(GO:0043776) cobalt-precorrin-7 C15-methyltransferase activity(GO:0043777) cobalt-precorrin-5B C1-methyltransferase activity(GO:0043780) cobalt-precorrin-3 C17-methyltransferase activity(GO:0043782) dimethylamine methyltransferase activity(GO:0043791) hydroxyneurosporene-O-methyltransferase activity(GO:0043803) tRNA (adenine-57, 58-N(1)-) methyltransferase activity(GO:0043827) methylamine-specific methylcobalamin:coenzyme M methyltransferase activity(GO:0043833) trimethylamine methyltransferase activity(GO:0043834) methanol-specific methylcobalamin:coenzyme M methyltransferase activity(GO:0043851) monomethylamine methyltransferase activity(GO:0043852) P-methyltransferase activity(GO:0051994) Se-methyltransferase activity(GO:0051995) 2-phytyl-1,4-naphthoquinone methyltransferase activity(GO:0052624) tRNA (uracil-2'-O-)-methyltransferase activity(GO:0052665) tRNA (cytosine-2'-O-)-methyltransferase activity(GO:0052666) phosphomethylethanolamine N-methyltransferase activity(GO:0052667) tRNA (cytosine-3-)-methyltransferase activity(GO:0052735) rRNA (cytosine-2'-O-)-methyltransferase activity(GO:0070677) rRNA (cytosine-N4-)-methyltransferase activity(GO:0071424) trihydroxyferuloyl spermidine O-methyltransferase activity(GO:0080012) |
| 0.1 | 1.5 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.4 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.8 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.5 | GO:0036442 | hydrogen-exporting ATPase activity(GO:0036442) ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 3.9 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.9 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 2.4 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 1.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 1.0 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 1.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 2.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.3 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 1.2 | GO:0018602 | sulfonate dioxygenase activity(GO:0000907) 2,4-dichlorophenoxyacetate alpha-ketoglutarate dioxygenase activity(GO:0018602) hypophosphite dioxygenase activity(GO:0034792) gibberellin 2-beta-dioxygenase activity(GO:0045543) C-19 gibberellin 2-beta-dioxygenase activity(GO:0052634) C-20 gibberellin 2-beta-dioxygenase activity(GO:0052635) |
| 0.0 | 0.7 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.0 | 0.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.2 | GO:0016004 | phospholipase activator activity(GO:0016004) lipase activator activity(GO:0060229) |
| 0.0 | 0.7 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 1.2 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.6 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 1.6 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 1.8 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.8 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 2.1 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.1 | GO:0070181 | mRNA 5'-UTR binding(GO:0048027) small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.6 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 7.3 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 2.8 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 0.3 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |