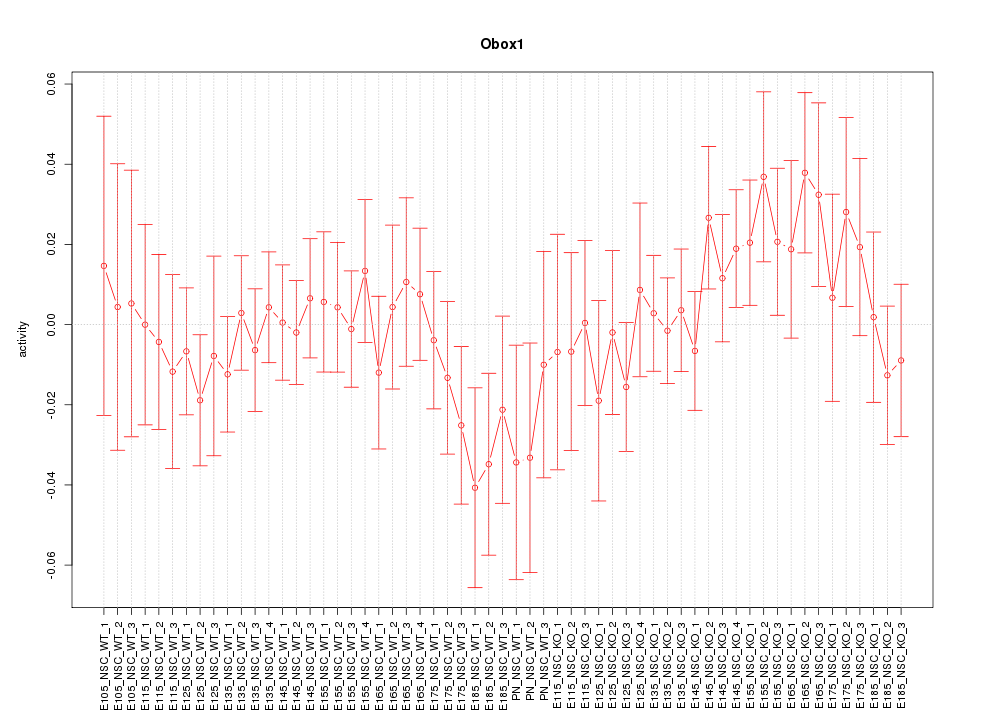
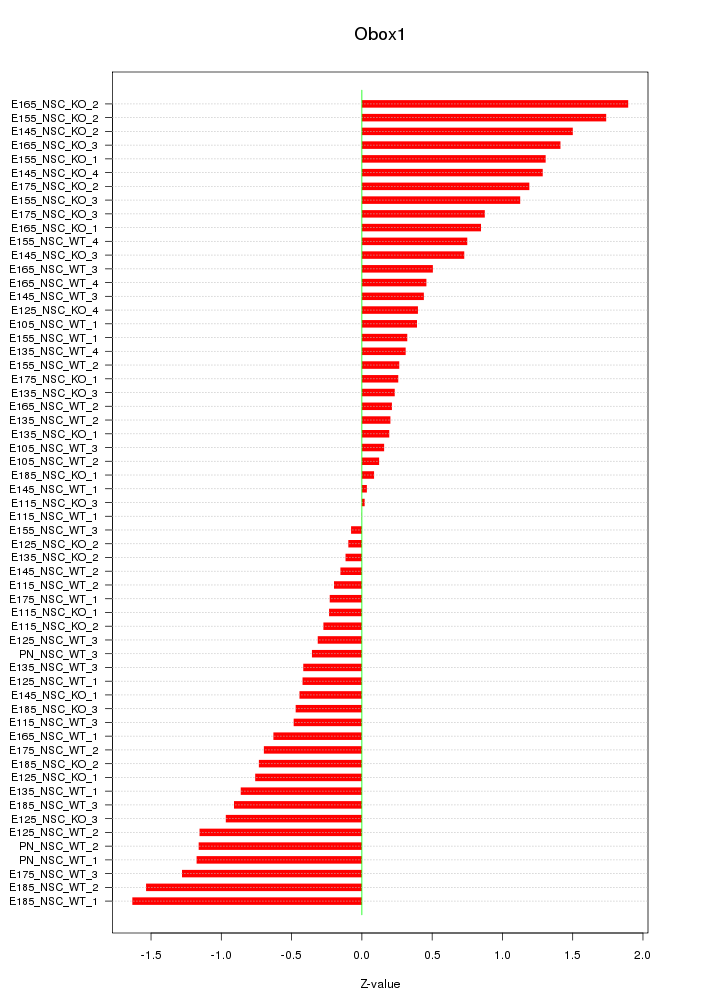
Motif ID: Obox1
Z-value: 0.800
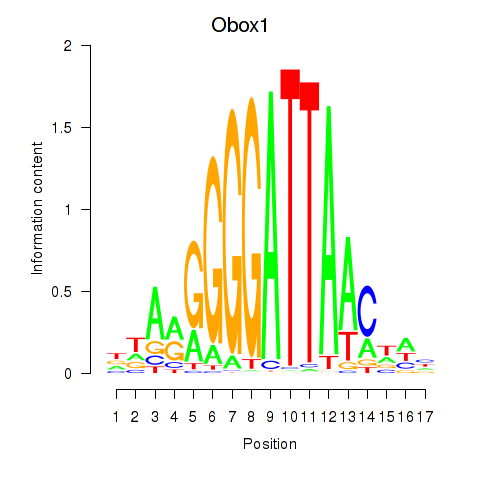
Transcription factors associated with Obox1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Obox1 | ENSMUSG00000054310.10 | Obox1 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0042822 | vitamin B6 metabolic process(GO:0042816) pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 1.0 | 2.9 | GO:0010958 | regulation of amino acid import(GO:0010958) regulation of L-arginine import(GO:0010963) |
| 0.9 | 3.5 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.9 | 13.8 | GO:0097688 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.6 | 4.1 | GO:0071361 | cellular response to zinc ion(GO:0071294) cellular response to ethanol(GO:0071361) |
| 0.5 | 1.6 | GO:0046104 | thymidine metabolic process(GO:0046104) |
| 0.5 | 0.5 | GO:0097503 | sialylation(GO:0097503) |
| 0.4 | 3.5 | GO:0034720 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) histone H3-K4 demethylation(GO:0034720) mammary duct terminal end bud growth(GO:0060763) |
| 0.4 | 1.6 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.4 | 7.2 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.4 | 1.1 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.4 | 3.6 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.3 | 1.1 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.3 | 3.7 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.2 | 1.5 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.2 | 0.7 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.2 | 1.2 | GO:0035247 | peptidyl-arginine omega-N-methylation(GO:0035247) histone H4-R3 methylation(GO:0043985) |
| 0.2 | 1.8 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.2 | 0.9 | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) |
| 0.2 | 1.3 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.2 | 0.8 | GO:0070942 | neutrophil mediated cytotoxicity(GO:0070942) neutrophil mediated killing of symbiont cell(GO:0070943) |
| 0.2 | 0.6 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.2 | 3.7 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.2 | 0.6 | GO:0031990 | mRNA export from nucleus in response to heat stress(GO:0031990) |
| 0.2 | 0.6 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.2 | 0.7 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.2 | 1.2 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.2 | 0.7 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.2 | 3.2 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.2 | 1.0 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.2 | 0.6 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.2 | 1.4 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.1 | 1.0 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.7 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.1 | 0.8 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.1 | 1.7 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 1.6 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 0.4 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.1 | 0.3 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.1 | 0.9 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 1.5 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.1 | 1.7 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.1 | 1.5 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 0.7 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.1 | 0.6 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.4 | GO:0032621 | interleukin-18 production(GO:0032621) positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.1 | 0.3 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.1 | 0.3 | GO:0046078 | pyrimidine deoxyribonucleotide catabolic process(GO:0009223) dUMP metabolic process(GO:0046078) |
| 0.1 | 0.7 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.1 | 0.9 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 0.9 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.1 | 2.4 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 0.9 | GO:0045324 | nuclear envelope reassembly(GO:0031468) late endosome to vacuole transport(GO:0045324) |
| 0.1 | 0.2 | GO:0052428 | modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.1 | 0.7 | GO:0030388 | fructose 6-phosphate metabolic process(GO:0006002) fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 1.9 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 1.2 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.1 | 3.2 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 0.3 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 0.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.6 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 0.6 | GO:2000010 | positive regulation of protein localization to cell surface(GO:2000010) |
| 0.1 | 2.2 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.1 | 0.9 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.1 | 4.7 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.1 | 0.4 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.7 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.0 | 0.2 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.0 | 0.2 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.0 | 0.2 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 4.8 | GO:0098792 | xenophagy(GO:0098792) |
| 0.0 | 0.2 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 1.6 | GO:0001782 | B cell homeostasis(GO:0001782) |
| 0.0 | 0.2 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.1 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.0 | 0.9 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.8 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.9 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.2 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.6 | GO:0097484 | dendrite extension(GO:0097484) |
| 0.0 | 0.1 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.0 | 0.4 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.7 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 1.4 | GO:0071806 | intracellular protein transmembrane transport(GO:0065002) protein transmembrane transport(GO:0071806) |
| 0.0 | 0.3 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 1.4 | GO:0055010 | ventricular cardiac muscle tissue morphogenesis(GO:0055010) |
| 0.0 | 1.0 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 1.0 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 1.0 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 3.7 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.6 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.6 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.4 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.7 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 1.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 6.4 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.2 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 1.0 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.2 | GO:0046036 | CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.0 | 2.1 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 1.0 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.5 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.7 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.7 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 6.1 | GO:0007186 | G-protein coupled receptor signaling pathway(GO:0007186) |
| 0.0 | 0.6 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.5 | GO:0035065 | regulation of histone acetylation(GO:0035065) |
| 0.0 | 0.1 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 10.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.5 | 3.6 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.5 | 1.5 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.3 | 3.5 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.3 | 1.0 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.3 | 3.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.3 | 1.4 | GO:0002177 | manchette(GO:0002177) |
| 0.2 | 0.7 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.2 | 8.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.2 | 0.7 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.2 | 1.6 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.2 | 0.8 | GO:0031523 | Myb complex(GO:0031523) |
| 0.1 | 1.4 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 1.4 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 1.2 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 3.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 3.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 0.9 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.7 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.9 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.7 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 2.9 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 1.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 0.7 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 1.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.9 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 0.6 | GO:0061700 | nuclear pore outer ring(GO:0031080) GATOR2 complex(GO:0061700) |
| 0.0 | 0.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.7 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.6 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.9 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.3 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 6.9 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 2.8 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 4.1 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 2.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.8 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.5 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:1990047 | spindle matrix(GO:1990047) |
| 0.0 | 1.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.4 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 1.0 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.1 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.3 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.3 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.1 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 1.0 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.7 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 0.5 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.9 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.7 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.4 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 2.1 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 1.3 | 4.0 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 1.3 | 10.3 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 1.2 | 3.5 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.7 | 2.9 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.7 | 3.5 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.5 | 3.6 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.5 | 1.5 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.5 | 2.9 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.5 | 8.1 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.4 | 1.6 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.4 | 1.6 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.3 | 1.0 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.3 | 1.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.3 | 2.4 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.3 | 0.8 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.2 | 7.2 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.2 | 1.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 0.7 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.2 | 1.4 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.2 | 2.9 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.2 | 3.7 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.2 | 0.4 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.2 | 0.8 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.2 | 0.7 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.2 | 0.7 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.2 | 1.5 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.2 | 0.6 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.2 | 1.7 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 4.9 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.1 | 0.9 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.6 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.7 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.1 | 1.6 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.1 | 0.4 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 1.0 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 1.8 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 2.8 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.6 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 0.9 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 1.3 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 1.1 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 0.9 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.1 | 0.9 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 2.9 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.4 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.1 | 0.6 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.8 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.4 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.1 | 0.3 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 0.1 | 0.6 | GO:0008251 | tRNA-specific adenosine deaminase activity(GO:0008251) |
| 0.1 | 0.6 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 2.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.3 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 0.2 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.1 | 0.2 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0005047 | signal recognition particle binding(GO:0005047) endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.7 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.9 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 1.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.9 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.2 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.0 | 1.0 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 2.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.5 | GO:0008932 | lytic endotransglycosylase activity(GO:0008932) |
| 0.0 | 0.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.7 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 1.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.7 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 13.1 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.6 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.8 | GO:0008748 | N-ethylmaleimide reductase activity(GO:0008748) reduced coenzyme F420 dehydrogenase activity(GO:0043738) sulfur oxygenase reductase activity(GO:0043826) malolactic enzyme activity(GO:0043883) epoxyqueuosine reductase activity(GO:0052693) |
| 0.0 | 0.2 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 5.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.2 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 1.0 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 0.6 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 0.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.9 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 1.2 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.4 | GO:0045182 | translation regulator activity(GO:0045182) |