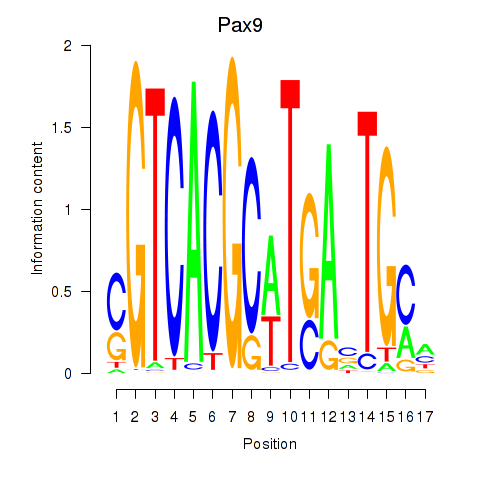
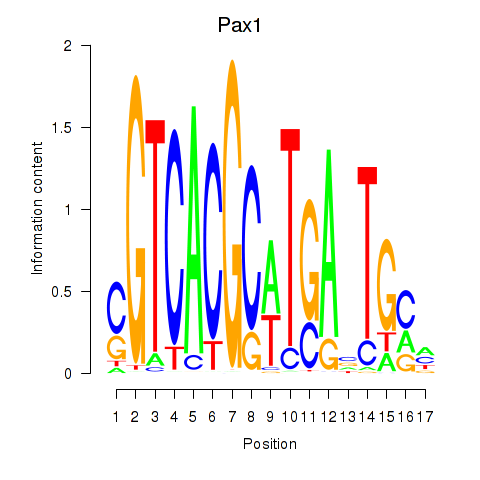
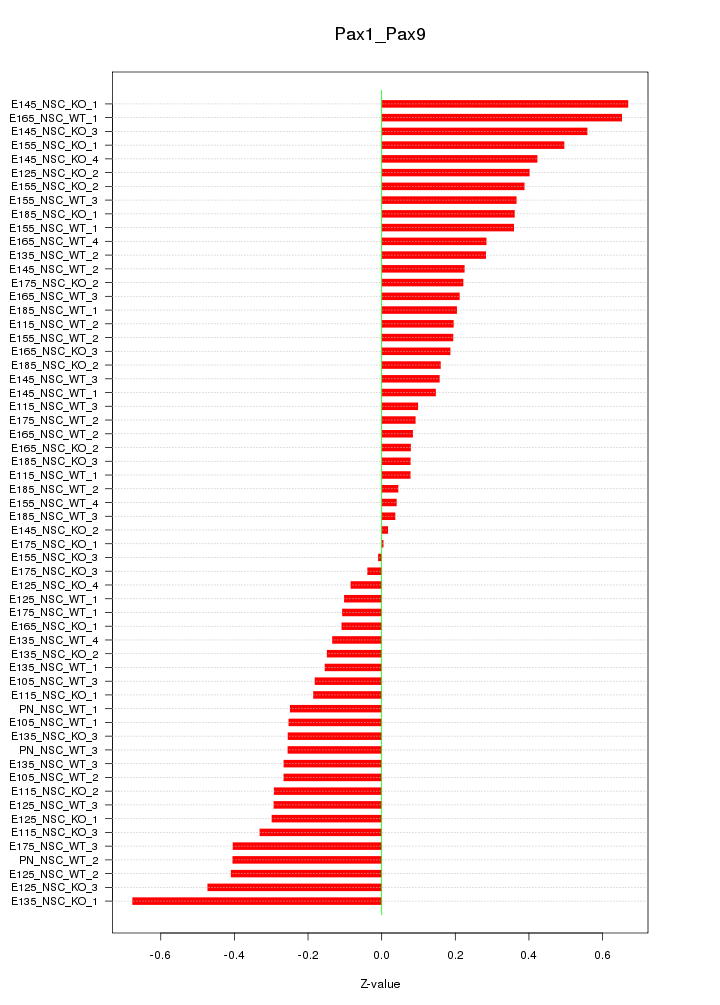
| 0.2 |
1.0 |
GO:0001011 |
transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 |
1.6 |
GO:0008599 |
protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.0 |
1.0 |
GO:0034930 |
N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) heparan sulfate 2-O-sulfotransferase activity(GO:0004394) HNK-1 sulfotransferase activity(GO:0016232) heparan sulfate 6-O-sulfotransferase activity(GO:0017095) trans-9R,10R-dihydrodiolphenanthrene sulfotransferase activity(GO:0018721) 1-phenanthrol sulfotransferase activity(GO:0018722) 3-phenanthrol sulfotransferase activity(GO:0018723) 4-phenanthrol sulfotransferase activity(GO:0018724) trans-3,4-dihydrodiolphenanthrene sulfotransferase activity(GO:0018725) 9-phenanthrol sulfotransferase activity(GO:0018726) 2-phenanthrol sulfotransferase activity(GO:0018727) phenanthrol sulfotransferase activity(GO:0019111) 1-hydroxypyrene sulfotransferase activity(GO:0034930) proteoglycan sulfotransferase activity(GO:0050698) cholesterol sulfotransferase activity(GO:0051922) hydroxyjasmonate sulfotransferase activity(GO:0080131) |
| 0.0 |
0.6 |
GO:0005104 |
fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 |
0.3 |
GO:0015215 |
nucleotide transmembrane transporter activity(GO:0015215) |
| 0.0 |
0.5 |
GO:0017091 |
AU-rich element binding(GO:0017091) |
| 0.0 |
0.4 |
GO:0000217 |
DNA secondary structure binding(GO:0000217) |
| 0.0 |
0.6 |
GO:0016538 |
cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |