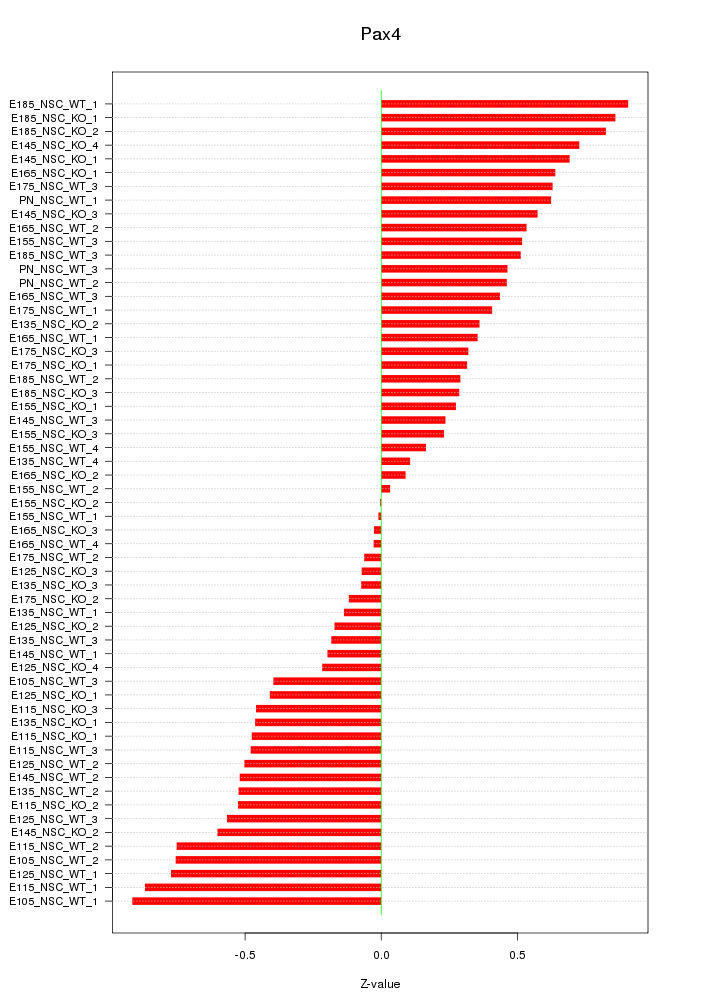
Motif ID: Pax4
Z-value: 0.482
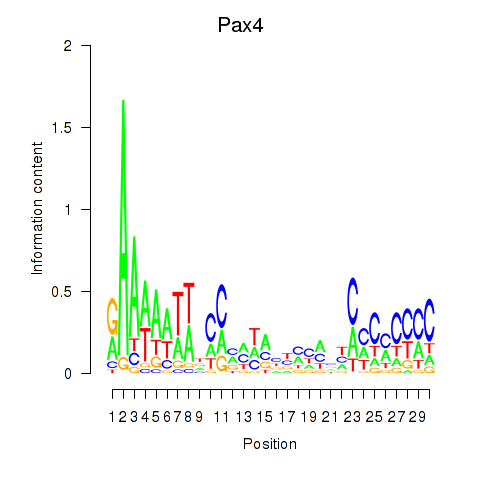
Transcription factors associated with Pax4:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Pax4 | ENSMUSG00000029706.9 | Pax4 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.9 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) |
| 0.9 | 3.4 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.8 | 3.1 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.6 | 2.9 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.5 | 1.5 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.3 | 4.7 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.3 | 1.8 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.3 | 3.5 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.3 | 2.0 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.3 | 2.8 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.3 | 1.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.2 | 1.5 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.2 | 1.9 | GO:0007379 | segment specification(GO:0007379) |
| 0.1 | 2.1 | GO:1902993 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.1 | 1.4 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.1 | 4.3 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 0.6 | GO:0002326 | B cell lineage commitment(GO:0002326) ectopic germ cell programmed cell death(GO:0035234) |
| 0.1 | 0.9 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.9 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 0.6 | GO:0097460 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) ferrous iron import into cell(GO:0097460) |
| 0.0 | 0.5 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 1.0 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 | 4.6 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.7 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.0 | 1.2 | GO:0090280 | positive regulation of calcium ion import(GO:0090280) |
| 0.0 | 0.7 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.2 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.5 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.3 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.4 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.5 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.1 | 2.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 1.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 2.0 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 0.6 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.1 | 1.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.6 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 0.8 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 7.5 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.6 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 1.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 1.0 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 4.4 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 1.8 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 7.7 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.3 | GO:0000788 | nuclear nucleosome(GO:0000788) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.8 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.3 | 1.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.2 | 0.6 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.2 | 3.1 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.2 | 3.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 1.8 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.6 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.1 | 0.5 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.9 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 0.9 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 3.4 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.7 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 1.0 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 1.4 | GO:0004120 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) photoreceptor cyclic-nucleotide phosphodiesterase activity(GO:0004120) 7,8-dihydro-D-neopterin 2',3'-cyclic phosphate phosphodiesterase activity(GO:0044688) inositol phosphosphingolipid phospholipase activity(GO:0052712) inositol phosphorylceramide phospholipase activity(GO:0052713) mannosyl-inositol phosphorylceramide phospholipase activity(GO:0052714) mannosyl-diinositol phosphorylceramide phospholipase activity(GO:0052715) |
| 0.0 | 2.9 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 2.1 | GO:0019210 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.0 | 2.0 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 1.0 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.3 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 5.0 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.8 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 1.2 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 1.5 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.8 | GO:0051219 | phosphoprotein binding(GO:0051219) |