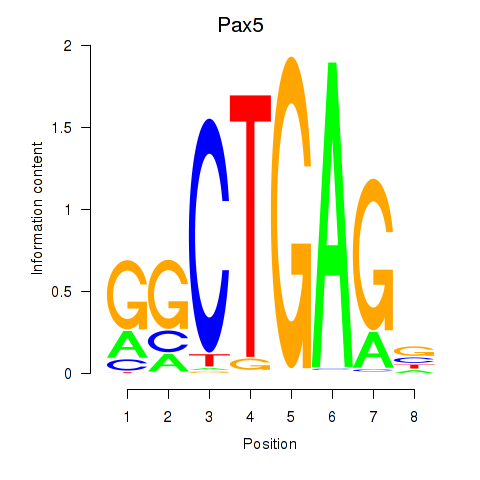
| 3.6 |
10.8 |
GO:1903279 |
regulation of calcium:sodium antiporter activity(GO:1903279) |
| 3.6 |
21.6 |
GO:0010624 |
regulation of Schwann cell proliferation(GO:0010624) |
| 3.6 |
10.7 |
GO:0014012 |
peripheral nervous system axon regeneration(GO:0014012) |
| 1.9 |
5.6 |
GO:0090425 |
hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 1.7 |
5.0 |
GO:0043465 |
regulation of fermentation(GO:0043465) negative regulation of fermentation(GO:1901003) |
| 1.6 |
6.5 |
GO:2000427 |
positive regulation of apoptotic cell clearance(GO:2000427) |
| 1.5 |
6.2 |
GO:0042938 |
dipeptide transport(GO:0042938) |
| 1.4 |
16.4 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 1.3 |
3.8 |
GO:0048686 |
regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 1.2 |
7.3 |
GO:0007217 |
tachykinin receptor signaling pathway(GO:0007217) |
| 1.2 |
12.1 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 1.2 |
4.8 |
GO:0001920 |
negative regulation of receptor recycling(GO:0001920) |
| 1.1 |
4.5 |
GO:0035617 |
stress granule disassembly(GO:0035617) |
| 1.1 |
3.4 |
GO:0061153 |
trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 1.1 |
4.4 |
GO:0006867 |
asparagine transport(GO:0006867) glutamine transport(GO:0006868) |
| 1.1 |
13.8 |
GO:0070842 |
aggresome assembly(GO:0070842) |
| 1.0 |
3.1 |
GO:2000111 |
positive regulation of macrophage apoptotic process(GO:2000111) |
| 1.0 |
6.1 |
GO:0033600 |
negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 1.0 |
3.0 |
GO:0036092 |
phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 1.0 |
3.0 |
GO:0010751 |
regulation of arginine metabolic process(GO:0000821) negative regulation of nitric oxide mediated signal transduction(GO:0010751) negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 1.0 |
1.0 |
GO:0035910 |
ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 1.0 |
4.0 |
GO:0019372 |
lipoxygenase pathway(GO:0019372) |
| 1.0 |
3.0 |
GO:0061743 |
motor learning(GO:0061743) |
| 0.9 |
0.9 |
GO:0018307 |
enzyme active site formation(GO:0018307) |
| 0.9 |
10.9 |
GO:0048484 |
enteric nervous system development(GO:0048484) |
| 0.9 |
2.7 |
GO:0003357 |
noradrenergic neuron differentiation(GO:0003357) |
| 0.9 |
19.0 |
GO:0015701 |
bicarbonate transport(GO:0015701) |
| 0.9 |
3.6 |
GO:0045113 |
regulation of integrin biosynthetic process(GO:0045113) |
| 0.9 |
4.3 |
GO:0050847 |
progesterone receptor signaling pathway(GO:0050847) |
| 0.8 |
11.7 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.8 |
2.5 |
GO:0060060 |
post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.8 |
0.8 |
GO:2000977 |
regulation of forebrain neuron differentiation(GO:2000977) |
| 0.8 |
4.0 |
GO:0051045 |
negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.7 |
2.2 |
GO:1900365 |
positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.7 |
2.2 |
GO:0016256 |
N-glycan processing to lysosome(GO:0016256) |
| 0.7 |
2.2 |
GO:0006553 |
lysine metabolic process(GO:0006553) |
| 0.7 |
4.4 |
GO:0048934 |
peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.7 |
5.0 |
GO:0045213 |
neurotransmitter receptor metabolic process(GO:0045213) |
| 0.7 |
2.1 |
GO:0001978 |
regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) |
| 0.7 |
2.0 |
GO:0010636 |
positive regulation of mitochondrial fusion(GO:0010636) |
| 0.6 |
3.1 |
GO:0038026 |
reelin-mediated signaling pathway(GO:0038026) |
| 0.6 |
1.9 |
GO:0019858 |
cytosine metabolic process(GO:0019858) |
| 0.6 |
2.5 |
GO:0035063 |
nuclear speck organization(GO:0035063) |
| 0.6 |
1.2 |
GO:1902548 |
negative regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902548) |
| 0.6 |
3.0 |
GO:0050862 |
positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.6 |
1.8 |
GO:0048162 |
preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.6 |
5.3 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.6 |
4.6 |
GO:0061635 |
regulation of protein complex stability(GO:0061635) |
| 0.6 |
4.0 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.6 |
2.2 |
GO:0010808 |
positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.6 |
5.6 |
GO:0030388 |
fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.6 |
2.8 |
GO:0034372 |
very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.5 |
1.6 |
GO:0042138 |
meiotic DNA double-strand break formation(GO:0042138) |
| 0.5 |
1.1 |
GO:0002182 |
cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.5 |
6.4 |
GO:0033008 |
positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.5 |
2.1 |
GO:0071394 |
cellular response to testosterone stimulus(GO:0071394) |
| 0.5 |
2.6 |
GO:0007262 |
STAT protein import into nucleus(GO:0007262) |
| 0.5 |
3.7 |
GO:0043553 |
negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.5 |
2.0 |
GO:0033540 |
fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.5 |
2.5 |
GO:0019236 |
response to pheromone(GO:0019236) |
| 0.5 |
2.5 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |
| 0.5 |
8.4 |
GO:0030204 |
chondroitin sulfate metabolic process(GO:0030204) |
| 0.5 |
2.0 |
GO:2000795 |
negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.5 |
1.5 |
GO:1904742 |
regulation of telomeric DNA binding(GO:1904742) |
| 0.5 |
1.4 |
GO:0031424 |
keratinization(GO:0031424) |
| 0.5 |
2.8 |
GO:1903056 |
regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.5 |
2.8 |
GO:1900246 |
positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.5 |
1.4 |
GO:0033566 |
gamma-tubulin complex localization(GO:0033566) |
| 0.5 |
3.7 |
GO:0048149 |
behavioral response to ethanol(GO:0048149) |
| 0.5 |
1.4 |
GO:0097051 |
establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.4 |
0.9 |
GO:2000468 |
regulation of peroxidase activity(GO:2000468) |
| 0.4 |
1.7 |
GO:1903031 |
regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.4 |
5.6 |
GO:0071257 |
cellular response to electrical stimulus(GO:0071257) |
| 0.4 |
8.4 |
GO:0048712 |
negative regulation of astrocyte differentiation(GO:0048712) |
| 0.4 |
1.3 |
GO:0021546 |
rhombomere development(GO:0021546) |
| 0.4 |
5.7 |
GO:0019243 |
methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.4 |
1.2 |
GO:0050975 |
sensory perception of touch(GO:0050975) |
| 0.4 |
2.7 |
GO:0030718 |
germ-line stem cell population maintenance(GO:0030718) |
| 0.4 |
6.2 |
GO:0015693 |
magnesium ion transport(GO:0015693) |
| 0.4 |
1.9 |
GO:0060762 |
regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.4 |
2.3 |
GO:0032261 |
purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.4 |
1.9 |
GO:0032237 |
activation of store-operated calcium channel activity(GO:0032237) |
| 0.4 |
1.1 |
GO:0044268 |
multicellular organismal protein metabolic process(GO:0044268) |
| 0.4 |
3.0 |
GO:0006620 |
posttranslational protein targeting to membrane(GO:0006620) |
| 0.4 |
1.8 |
GO:0060075 |
regulation of resting membrane potential(GO:0060075) |
| 0.4 |
1.5 |
GO:0097167 |
circadian regulation of translation(GO:0097167) |
| 0.4 |
1.1 |
GO:0032058 |
positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.4 |
1.8 |
GO:0014054 |
positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.4 |
4.4 |
GO:1902993 |
positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.4 |
7.3 |
GO:0032757 |
positive regulation of interleukin-8 production(GO:0032757) |
| 0.4 |
1.1 |
GO:0010796 |
regulation of multivesicular body size(GO:0010796) |
| 0.4 |
1.1 |
GO:0072554 |
blood vessel lumenization(GO:0072554) |
| 0.4 |
2.1 |
GO:0009786 |
regulation of asymmetric cell division(GO:0009786) |
| 0.4 |
4.6 |
GO:0042711 |
maternal behavior(GO:0042711) |
| 0.3 |
1.4 |
GO:0043974 |
histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.3 |
4.9 |
GO:0072010 |
renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.3 |
1.0 |
GO:1905154 |
negative regulation of tumor necrosis factor secretion(GO:1904468) negative regulation of membrane invagination(GO:1905154) |
| 0.3 |
1.7 |
GO:2000574 |
regulation of microtubule motor activity(GO:2000574) |
| 0.3 |
0.7 |
GO:0031394 |
positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.3 |
1.4 |
GO:1900740 |
membrane fission(GO:0090148) regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) regulation of membrane tubulation(GO:1903525) |
| 0.3 |
2.0 |
GO:0045743 |
positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.3 |
1.7 |
GO:0060178 |
regulation of exocyst localization(GO:0060178) |
| 0.3 |
1.0 |
GO:0034035 |
purine ribonucleoside bisphosphate metabolic process(GO:0034035) |
| 0.3 |
2.0 |
GO:0015879 |
carnitine transport(GO:0015879) |
| 0.3 |
1.6 |
GO:0035590 |
purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.3 |
0.9 |
GO:0046368 |
GDP-L-fucose metabolic process(GO:0046368) |
| 0.3 |
1.6 |
GO:0060278 |
regulation of ovulation(GO:0060278) |
| 0.3 |
7.1 |
GO:0051894 |
positive regulation of focal adhesion assembly(GO:0051894) |
| 0.3 |
0.9 |
GO:2000686 |
regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.3 |
2.4 |
GO:2000809 |
neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.3 |
4.7 |
GO:0032060 |
bleb assembly(GO:0032060) |
| 0.3 |
0.9 |
GO:0071929 |
alpha-tubulin acetylation(GO:0071929) |
| 0.3 |
2.3 |
GO:0033564 |
anterior/posterior axon guidance(GO:0033564) |
| 0.3 |
2.6 |
GO:0014049 |
positive regulation of glutamate secretion(GO:0014049) |
| 0.3 |
4.1 |
GO:0043508 |
negative regulation of JUN kinase activity(GO:0043508) |
| 0.3 |
1.7 |
GO:0007197 |
adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.3 |
0.9 |
GO:0019401 |
alditol biosynthetic process(GO:0019401) |
| 0.3 |
0.9 |
GO:0032916 |
positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 0.3 |
4.0 |
GO:0033866 |
coenzyme A biosynthetic process(GO:0015937) nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.3 |
1.7 |
GO:0034983 |
peptidyl-lysine deacetylation(GO:0034983) |
| 0.3 |
2.0 |
GO:1904217 |
regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.3 |
1.4 |
GO:1900095 |
regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.3 |
1.4 |
GO:0001923 |
B-1 B cell differentiation(GO:0001923) |
| 0.3 |
1.1 |
GO:0048245 |
eosinophil chemotaxis(GO:0048245) |
| 0.3 |
1.3 |
GO:0000098 |
sulfur amino acid catabolic process(GO:0000098) |
| 0.3 |
3.6 |
GO:0045579 |
positive regulation of B cell differentiation(GO:0045579) |
| 0.3 |
1.8 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.3 |
2.9 |
GO:0031936 |
negative regulation of chromatin silencing(GO:0031936) |
| 0.3 |
1.8 |
GO:0036120 |
cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.3 |
5.4 |
GO:0048268 |
clathrin coat assembly(GO:0048268) |
| 0.3 |
1.8 |
GO:0070447 |
positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.3 |
6.8 |
GO:0042462 |
eye photoreceptor cell development(GO:0042462) |
| 0.2 |
0.7 |
GO:0097151 |
positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 0.2 |
2.2 |
GO:0015868 |
purine ribonucleotide transport(GO:0015868) |
| 0.2 |
1.2 |
GO:0045541 |
negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.2 |
1.2 |
GO:2000587 |
regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.2 |
0.7 |
GO:0048633 |
positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.2 |
4.7 |
GO:0030033 |
microvillus assembly(GO:0030033) |
| 0.2 |
0.7 |
GO:0060745 |
mammary gland branching involved in pregnancy(GO:0060745) epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) branch elongation involved in mammary gland duct branching(GO:0060751) |
| 0.2 |
1.6 |
GO:0002043 |
blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.2 |
1.2 |
GO:0036151 |
phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.2 |
5.7 |
GO:0097120 |
receptor localization to synapse(GO:0097120) |
| 0.2 |
2.0 |
GO:0036158 |
outer dynein arm assembly(GO:0036158) |
| 0.2 |
3.6 |
GO:0014002 |
astrocyte development(GO:0014002) |
| 0.2 |
0.7 |
GO:0061537 |
glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.2 |
0.9 |
GO:0035624 |
receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.2 |
0.4 |
GO:1903903 |
regulation of establishment of T cell polarity(GO:1903903) |
| 0.2 |
2.4 |
GO:0010457 |
centriole-centriole cohesion(GO:0010457) |
| 0.2 |
1.7 |
GO:0035095 |
behavioral response to nicotine(GO:0035095) |
| 0.2 |
0.4 |
GO:2000393 |
negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.2 |
3.9 |
GO:0045475 |
locomotor rhythm(GO:0045475) |
| 0.2 |
16.8 |
GO:0010811 |
positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.2 |
2.0 |
GO:0097421 |
liver regeneration(GO:0097421) |
| 0.2 |
0.6 |
GO:2000813 |
negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.2 |
1.8 |
GO:0010606 |
positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.2 |
2.0 |
GO:1904153 |
negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.2 |
1.4 |
GO:0090336 |
positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.2 |
4.6 |
GO:0045662 |
negative regulation of myoblast differentiation(GO:0045662) |
| 0.2 |
1.0 |
GO:0010730 |
negative regulation of hydrogen peroxide metabolic process(GO:0010727) negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.2 |
5.0 |
GO:0007616 |
long-term memory(GO:0007616) |
| 0.2 |
2.2 |
GO:0019886 |
antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.2 |
1.2 |
GO:0001302 |
replicative cell aging(GO:0001302) |
| 0.2 |
0.2 |
GO:1900242 |
regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.2 |
4.7 |
GO:0046325 |
negative regulation of glucose import(GO:0046325) |
| 0.2 |
1.1 |
GO:0016557 |
peroxisome membrane biogenesis(GO:0016557) |
| 0.2 |
2.2 |
GO:0060213 |
regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.2 |
1.3 |
GO:0060080 |
inhibitory postsynaptic potential(GO:0060080) |
| 0.2 |
1.3 |
GO:0060081 |
membrane hyperpolarization(GO:0060081) |
| 0.2 |
1.5 |
GO:2000781 |
positive regulation of double-strand break repair(GO:2000781) |
| 0.2 |
5.7 |
GO:0007200 |
phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.2 |
5.9 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.2 |
1.4 |
GO:0016198 |
axon choice point recognition(GO:0016198) |
| 0.2 |
0.9 |
GO:1904936 |
cerebral cortex GABAergic interneuron migration(GO:0021853) cerebral cortex GABAergic interneuron development(GO:0021894) interneuron migration(GO:1904936) |
| 0.2 |
1.6 |
GO:0061088 |
sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.2 |
3.0 |
GO:1903861 |
regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.2 |
0.4 |
GO:2000224 |
testosterone biosynthetic process(GO:0061370) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.2 |
0.9 |
GO:0036006 |
response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.2 |
1.0 |
GO:0044829 |
positive regulation by host of viral genome replication(GO:0044829) |
| 0.2 |
1.4 |
GO:0019673 |
dolichol metabolic process(GO:0019348) GDP-mannose metabolic process(GO:0019673) |
| 0.2 |
4.3 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 |
0.5 |
GO:1904177 |
regulation of adipose tissue development(GO:1904177) |
| 0.2 |
0.7 |
GO:0046543 |
development of secondary female sexual characteristics(GO:0046543) |
| 0.2 |
0.2 |
GO:0042713 |
sperm ejaculation(GO:0042713) |
| 0.2 |
2.2 |
GO:1901844 |
regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.2 |
0.5 |
GO:0043084 |
penile erection(GO:0043084) |
| 0.2 |
0.7 |
GO:0051189 |
Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.2 |
1.3 |
GO:0021902 |
commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.2 |
0.3 |
GO:1902174 |
positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.2 |
1.2 |
GO:0035024 |
negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 |
0.7 |
GO:0019695 |
choline metabolic process(GO:0019695) |
| 0.1 |
0.7 |
GO:0090315 |
negative regulation of protein targeting to membrane(GO:0090315) |
| 0.1 |
0.7 |
GO:1901843 |
positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.1 |
0.7 |
GO:0035093 |
spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 |
0.3 |
GO:0061419 |
connective tissue replacement involved in inflammatory response wound healing(GO:0002248) positive regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061419) connective tissue replacement(GO:0097709) |
| 0.1 |
1.9 |
GO:0009072 |
aromatic amino acid family metabolic process(GO:0009072) |
| 0.1 |
0.3 |
GO:0035106 |
operant conditioning(GO:0035106) |
| 0.1 |
0.4 |
GO:0048211 |
Golgi vesicle docking(GO:0048211) |
| 0.1 |
0.8 |
GO:0043353 |
enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 |
0.4 |
GO:1901491 |
negative regulation of lymphangiogenesis(GO:1901491) |
| 0.1 |
0.6 |
GO:0033700 |
phospholipid efflux(GO:0033700) |
| 0.1 |
0.8 |
GO:1901897 |
regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.1 |
1.8 |
GO:0060394 |
negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 |
0.4 |
GO:0034729 |
histone H3-K79 methylation(GO:0034729) |
| 0.1 |
5.4 |
GO:0045600 |
positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 |
2.2 |
GO:0034643 |
establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.1 |
8.6 |
GO:0010923 |
negative regulation of phosphatase activity(GO:0010923) |
| 0.1 |
0.6 |
GO:0032929 |
negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 |
1.7 |
GO:0035563 |
positive regulation of chromatin binding(GO:0035563) |
| 0.1 |
1.0 |
GO:0007379 |
segment specification(GO:0007379) |
| 0.1 |
1.6 |
GO:0006268 |
DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 |
0.7 |
GO:0032483 |
regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 |
0.5 |
GO:0010961 |
cellular magnesium ion homeostasis(GO:0010961) |
| 0.1 |
0.5 |
GO:0098543 |
detection of bacterium(GO:0016045) detection of other organism(GO:0098543) |
| 0.1 |
1.0 |
GO:0007216 |
G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 |
0.6 |
GO:0046208 |
spermine catabolic process(GO:0046208) |
| 0.1 |
0.6 |
GO:0071712 |
ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 |
0.5 |
GO:0034975 |
protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 |
1.9 |
GO:0045760 |
positive regulation of action potential(GO:0045760) |
| 0.1 |
0.5 |
GO:0072321 |
chaperone-mediated protein transport(GO:0072321) |
| 0.1 |
2.4 |
GO:2000398 |
regulation of T cell differentiation in thymus(GO:0033081) regulation of thymocyte aggregation(GO:2000398) |
| 0.1 |
8.2 |
GO:0051592 |
response to calcium ion(GO:0051592) |
| 0.1 |
7.7 |
GO:0007601 |
visual perception(GO:0007601) |
| 0.1 |
1.3 |
GO:0010803 |
regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.1 |
0.4 |
GO:0032815 |
negative regulation of natural killer cell activation(GO:0032815) |
| 0.1 |
0.3 |
GO:0072189 |
ureter development(GO:0072189) |
| 0.1 |
4.2 |
GO:0048843 |
negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.1 |
0.6 |
GO:1903887 |
motile primary cilium assembly(GO:1903887) |
| 0.1 |
2.1 |
GO:0010800 |
positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.1 |
0.2 |
GO:0021826 |
substrate-independent telencephalic tangential migration(GO:0021826) substrate-independent telencephalic tangential interneuron migration(GO:0021843) |
| 0.1 |
0.5 |
GO:0038018 |
Wnt receptor catabolic process(GO:0038018) |
| 0.1 |
0.8 |
GO:0010960 |
magnesium ion homeostasis(GO:0010960) |
| 0.1 |
0.3 |
GO:0007354 |
zygotic determination of anterior/posterior axis, embryo(GO:0007354) |
| 0.1 |
1.2 |
GO:0045815 |
positive regulation of gene expression, epigenetic(GO:0045815) |
| 0.1 |
2.2 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.1 |
2.6 |
GO:0015804 |
neutral amino acid transport(GO:0015804) |
| 0.1 |
1.0 |
GO:0006892 |
post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.1 |
2.4 |
GO:0001523 |
retinoid metabolic process(GO:0001523) |
| 0.1 |
1.8 |
GO:0007340 |
acrosome reaction(GO:0007340) |
| 0.1 |
0.6 |
GO:0045162 |
clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 |
0.4 |
GO:0033136 |
serine phosphorylation of STAT3 protein(GO:0033136) serine phosphorylation of STAT protein(GO:0042501) |
| 0.1 |
1.0 |
GO:0090084 |
negative regulation of inclusion body assembly(GO:0090084) |
| 0.1 |
2.9 |
GO:0070979 |
protein K11-linked ubiquitination(GO:0070979) |
| 0.1 |
0.4 |
GO:2000651 |
positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 |
1.9 |
GO:0030212 |
hyaluronan metabolic process(GO:0030212) |
| 0.1 |
0.5 |
GO:0018125 |
peptidyl-cysteine methylation(GO:0018125) |
| 0.1 |
0.7 |
GO:0051926 |
negative regulation of calcium ion transport(GO:0051926) |
| 0.1 |
0.4 |
GO:2000051 |
negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 |
3.2 |
GO:0030514 |
negative regulation of BMP signaling pathway(GO:0030514) |
| 0.1 |
0.6 |
GO:1902035 |
positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.1 |
4.0 |
GO:0048278 |
vesicle docking(GO:0048278) |
| 0.1 |
1.0 |
GO:0034773 |
histone H4-K20 trimethylation(GO:0034773) |
| 0.1 |
1.2 |
GO:0097237 |
cellular response to toxic substance(GO:0097237) |
| 0.1 |
1.4 |
GO:0060009 |
Sertoli cell development(GO:0060009) |
| 0.1 |
1.0 |
GO:0043374 |
CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 |
1.2 |
GO:1903779 |
regulation of cardiac conduction(GO:1903779) |
| 0.1 |
0.3 |
GO:2000598 |
regulation of cyclin catabolic process(GO:2000598) negative regulation of cyclin catabolic process(GO:2000599) |
| 0.1 |
2.3 |
GO:0007218 |
neuropeptide signaling pathway(GO:0007218) |
| 0.1 |
2.1 |
GO:0046854 |
phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 |
3.0 |
GO:0048260 |
positive regulation of receptor-mediated endocytosis(GO:0048260) |
| 0.1 |
0.3 |
GO:0034393 |
positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.1 |
0.2 |
GO:0042756 |
drinking behavior(GO:0042756) |
| 0.1 |
0.9 |
GO:0070296 |
sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.1 |
2.0 |
GO:0086010 |
membrane depolarization during action potential(GO:0086010) |
| 0.1 |
0.5 |
GO:0048251 |
elastic fiber assembly(GO:0048251) |
| 0.1 |
1.1 |
GO:0031297 |
replication fork processing(GO:0031297) |
| 0.1 |
0.2 |
GO:0010566 |
regulation of ketone biosynthetic process(GO:0010566) |
| 0.1 |
2.7 |
GO:0035690 |
cellular response to drug(GO:0035690) |
| 0.1 |
5.4 |
GO:0043149 |
contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.1 |
0.1 |
GO:0071218 |
cellular response to misfolded protein(GO:0071218) |
| 0.1 |
0.8 |
GO:0045939 |
negative regulation of steroid biosynthetic process(GO:0010894) negative regulation of steroid metabolic process(GO:0045939) |
| 0.1 |
0.4 |
GO:0038030 |
non-canonical Wnt signaling pathway via MAPK cascade(GO:0038030) non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.1 |
8.1 |
GO:0006813 |
potassium ion transport(GO:0006813) |
| 0.1 |
0.6 |
GO:0001541 |
ovarian follicle development(GO:0001541) |
| 0.1 |
0.3 |
GO:0010668 |
ectodermal cell differentiation(GO:0010668) |
| 0.1 |
0.7 |
GO:0070734 |
histone H3-K27 methylation(GO:0070734) |
| 0.1 |
0.1 |
GO:0043652 |
engulfment of apoptotic cell(GO:0043652) |
| 0.1 |
1.2 |
GO:0045453 |
bone resorption(GO:0045453) |
| 0.1 |
1.9 |
GO:0015807 |
L-amino acid transport(GO:0015807) |
| 0.1 |
0.2 |
GO:1901534 |
positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.1 |
1.3 |
GO:0097352 |
autophagosome maturation(GO:0097352) |
| 0.1 |
1.0 |
GO:0032410 |
negative regulation of transporter activity(GO:0032410) |
| 0.1 |
0.1 |
GO:1904192 |
cholangiocyte apoptotic process(GO:1902488) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 0.1 |
1.4 |
GO:0007020 |
microtubule nucleation(GO:0007020) |
| 0.1 |
0.4 |
GO:1901385 |
regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 |
0.3 |
GO:0097119 |
postsynaptic density protein 95 clustering(GO:0097119) |
| 0.1 |
0.5 |
GO:0050982 |
detection of mechanical stimulus(GO:0050982) |
| 0.1 |
0.3 |
GO:0043314 |
negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) negative regulation of neutrophil activation(GO:1902564) |
| 0.1 |
0.6 |
GO:0014823 |
response to activity(GO:0014823) |
| 0.1 |
1.8 |
GO:0034605 |
cellular response to heat(GO:0034605) |
| 0.1 |
1.3 |
GO:0018345 |
protein palmitoylation(GO:0018345) |
| 0.1 |
0.6 |
GO:0030866 |
cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 |
2.7 |
GO:0007338 |
single fertilization(GO:0007338) |
| 0.0 |
7.0 |
GO:0008360 |
regulation of cell shape(GO:0008360) |
| 0.0 |
2.7 |
GO:0030968 |
endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 |
0.9 |
GO:0030890 |
positive regulation of B cell proliferation(GO:0030890) |
| 0.0 |
0.3 |
GO:0090218 |
positive regulation of lipid kinase activity(GO:0090218) |
| 0.0 |
4.6 |
GO:0007050 |
cell cycle arrest(GO:0007050) |
| 0.0 |
1.2 |
GO:0032007 |
negative regulation of TOR signaling(GO:0032007) |
| 0.0 |
0.4 |
GO:0045217 |
cell-cell junction maintenance(GO:0045217) |
| 0.0 |
0.9 |
GO:0044804 |
nucleophagy(GO:0044804) |
| 0.0 |
0.2 |
GO:2001197 |
positive regulation of extracellular matrix assembly(GO:1901203) regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 |
0.5 |
GO:0030970 |
retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 |
0.2 |
GO:0032957 |
inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 |
0.1 |
GO:0051788 |
response to misfolded protein(GO:0051788) |
| 0.0 |
0.2 |
GO:0045896 |
regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 |
1.6 |
GO:0050830 |
defense response to Gram-positive bacterium(GO:0050830) |
| 0.0 |
0.1 |
GO:0050882 |
voluntary musculoskeletal movement(GO:0050882) |
| 0.0 |
0.2 |
GO:0033184 |
positive regulation of histone ubiquitination(GO:0033184) |
| 0.0 |
0.3 |
GO:0010763 |
positive regulation of fibroblast migration(GO:0010763) |
| 0.0 |
1.0 |
GO:0061001 |
regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 |
0.1 |
GO:0070886 |
positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 |
0.4 |
GO:0090205 |
positive regulation of cholesterol biosynthetic process(GO:0045542) positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.0 |
0.2 |
GO:1901673 |
regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 |
2.3 |
GO:0009636 |
response to toxic substance(GO:0009636) |
| 0.0 |
3.2 |
GO:0035023 |
regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 |
0.9 |
GO:0034314 |
Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 |
1.5 |
GO:0017157 |
regulation of exocytosis(GO:0017157) |
| 0.0 |
0.1 |
GO:0060179 |
male mating behavior(GO:0060179) |
| 0.0 |
0.3 |
GO:0006449 |
regulation of translational termination(GO:0006449) |
| 0.0 |
0.3 |
GO:0007168 |
receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 |
0.3 |
GO:0060445 |
branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 |
0.2 |
GO:0001980 |
regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.0 |
0.4 |
GO:0070816 |
phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 |
0.1 |
GO:0070966 |
nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 |
0.2 |
GO:0033234 |
negative regulation of protein sumoylation(GO:0033234) |
| 0.0 |
0.1 |
GO:0000290 |
deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 |
0.5 |
GO:0021681 |
cerebellar granular layer development(GO:0021681) |
| 0.0 |
0.5 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.0 |
1.3 |
GO:0060976 |
coronary vasculature development(GO:0060976) |
| 0.0 |
1.5 |
GO:0010951 |
negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 |
0.7 |
GO:0046785 |
microtubule polymerization(GO:0046785) |
| 0.0 |
0.1 |
GO:1903300 |
negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 |
0.5 |
GO:0006111 |
regulation of gluconeogenesis(GO:0006111) |
| 0.0 |
0.8 |
GO:0051897 |
positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 |
0.6 |
GO:0097006 |
regulation of plasma lipoprotein particle levels(GO:0097006) |
| 0.0 |
1.7 |
GO:0001505 |
regulation of neurotransmitter levels(GO:0001505) |
| 0.0 |
0.1 |
GO:0033364 |
mast cell secretory granule organization(GO:0033364) |
| 0.0 |
0.4 |
GO:2000785 |
regulation of autophagosome assembly(GO:2000785) |
| 0.0 |
0.3 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 |
0.1 |
GO:0035020 |
regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 |
0.3 |
GO:0016486 |
peptide hormone processing(GO:0016486) |
| 0.0 |
0.1 |
GO:0032484 |
Ral protein signal transduction(GO:0032484) |
| 0.0 |
0.1 |
GO:0018344 |
protein geranylgeranylation(GO:0018344) |
| 0.0 |
0.7 |
GO:0006289 |
nucleotide-excision repair(GO:0006289) |
| 0.0 |
0.4 |
GO:0017145 |
stem cell division(GO:0017145) |
| 0.0 |
0.2 |
GO:0061036 |
positive regulation of cartilage development(GO:0061036) |
| 0.0 |
0.1 |
GO:0014829 |
vascular smooth muscle contraction(GO:0014829) |