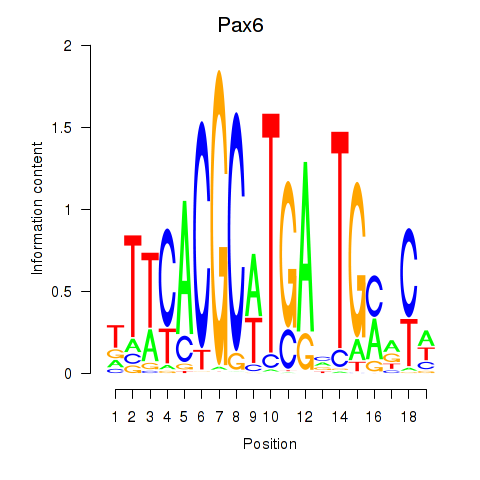
Motif ID: Pax6
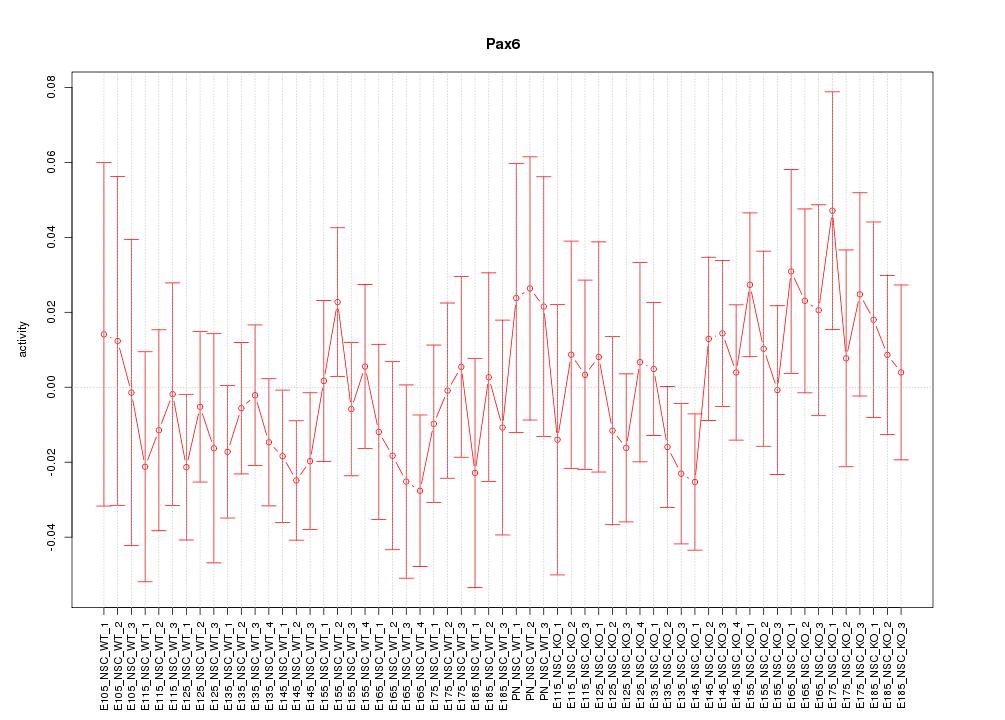
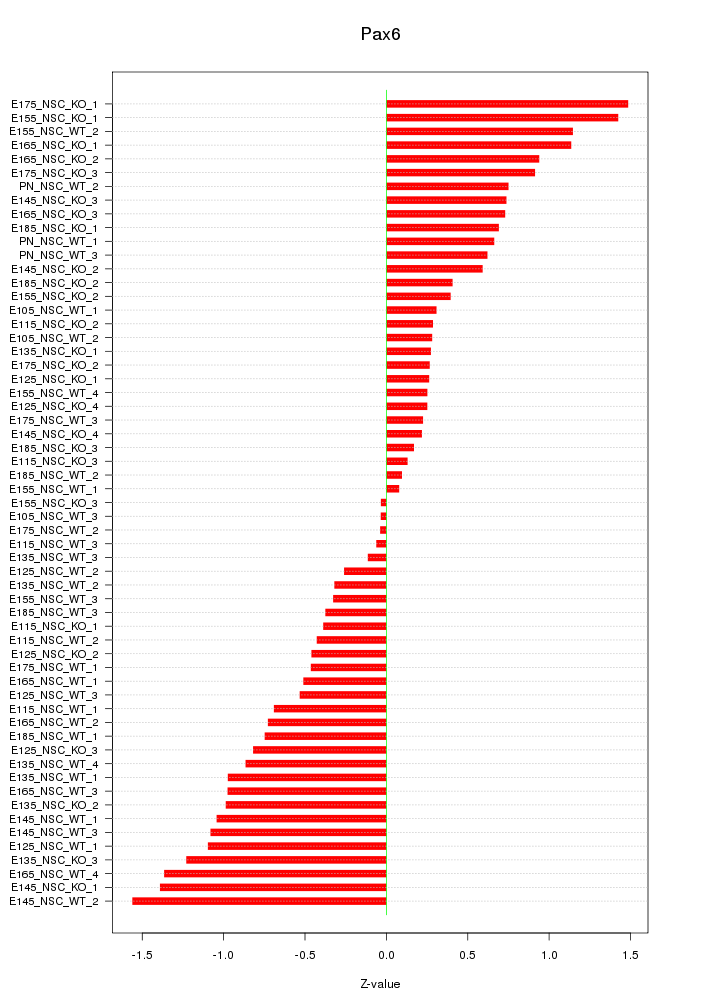
Z-value: 0.734
Transcription factors associated with Pax6:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Pax6 | ENSMUSG00000027168.15 | Pax6 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pax6 | mm10_v2_chr2_+_105668888_105668914 | -0.52 | 2.5e-05 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.6 | 3.2 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.5 | 1.0 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.5 | 2.4 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.4 | 1.7 | GO:0010040 | response to iron(II) ion(GO:0010040) positive regulation of hydrogen peroxide metabolic process(GO:0010726) negative regulation of synaptic transmission, dopaminergic(GO:0032227) negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) cellular response to copper ion(GO:0071280) regulation of peroxidase activity(GO:2000468) |
| 0.4 | 1.5 | GO:0098912 | smooth muscle contraction involved in micturition(GO:0060083) membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.3 | 1.0 | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) |
| 0.3 | 1.1 | GO:0061724 | lipophagy(GO:0061724) |
| 0.3 | 0.8 | GO:0097274 | urea homeostasis(GO:0097274) |
| 0.2 | 0.7 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.1 | 1.0 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.1 | 2.1 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.1 | 0.8 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.4 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.1 | 0.6 | GO:0051661 | maintenance of centrosome location(GO:0051661) maintenance of Golgi location(GO:0051684) |
| 0.1 | 3.5 | GO:0006760 | folic acid-containing compound metabolic process(GO:0006760) |
| 0.1 | 0.8 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.9 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.1 | 1.3 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.1 | 1.8 | GO:0071624 | positive regulation of granulocyte chemotaxis(GO:0071624) positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.1 | 0.8 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 0.2 | GO:0035864 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.1 | 0.1 | GO:1990705 | cholangiocyte proliferation(GO:1990705) |
| 0.1 | 5.4 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 4.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.9 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.1 | 0.3 | GO:0009098 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.1 | 0.7 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.0 | 1.5 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.2 | GO:2000258 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 0.2 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.0 | 2.2 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 0.2 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.2 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.3 | GO:0032757 | positive regulation of interleukin-8 production(GO:0032757) |
| 0.0 | 1.1 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.0 | 1.4 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.2 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.0 | GO:0007603 | phototransduction, visible light(GO:0007603) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.4 | 1.7 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.3 | 2.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.2 | 2.4 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.2 | 1.5 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.6 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 0.8 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 1.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 0.7 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.1 | 0.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.6 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.1 | 5.6 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.6 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.2 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.7 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 1.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 1.0 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 1.9 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.2 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.4 | GO:0031902 | late endosome membrane(GO:0031902) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.7 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.4 | 3.2 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.3 | 1.0 | GO:0004942 | anaphylatoxin receptor activity(GO:0004942) |
| 0.3 | 4.3 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.3 | 0.8 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.3 | 2.4 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.2 | 3.5 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.2 | 1.3 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.2 | 0.6 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) |
| 0.2 | 1.8 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.2 | 2.9 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.2 | 1.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 2.1 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.1 | 2.2 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.7 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 0.3 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.4 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 1.6 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 1.7 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.8 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.4 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 6.2 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |