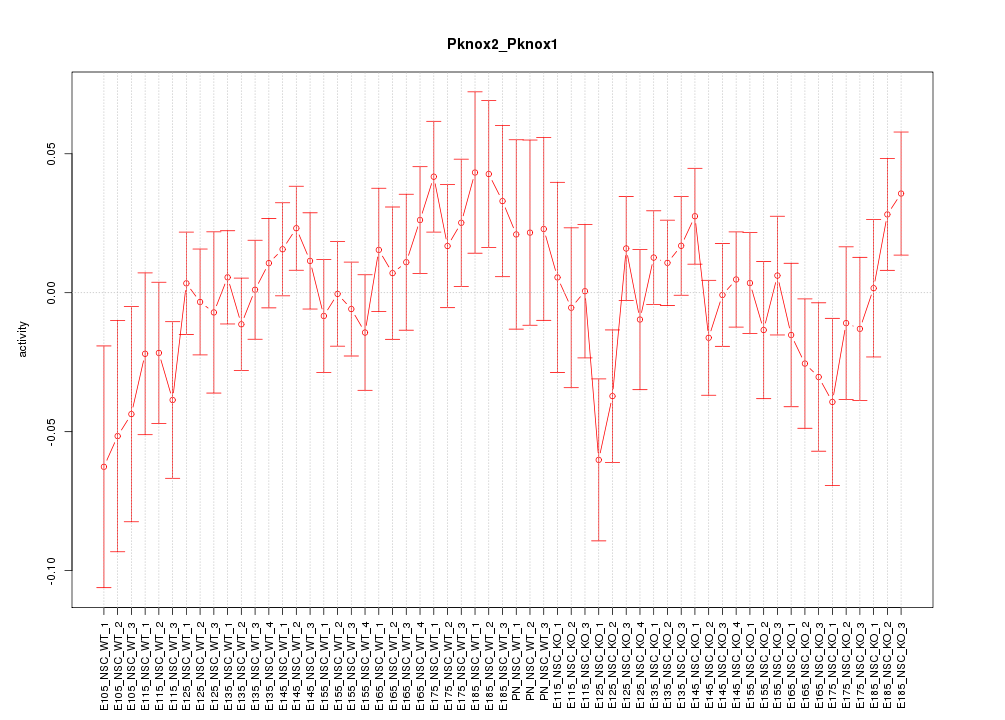
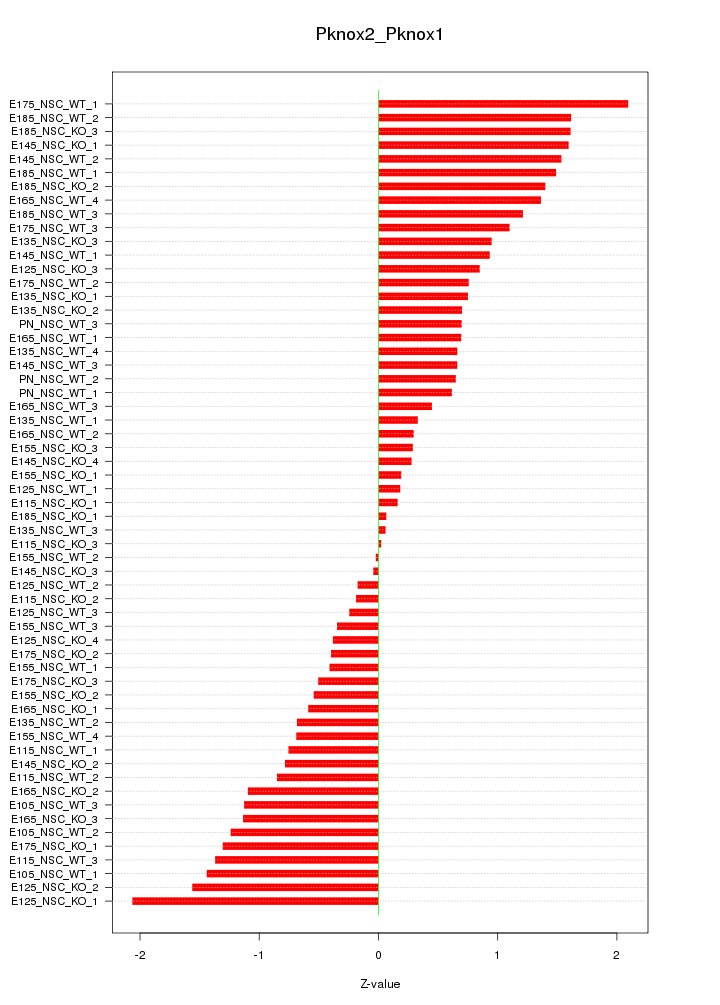
Motif ID: Pknox2_Pknox1
Z-value: 0.946
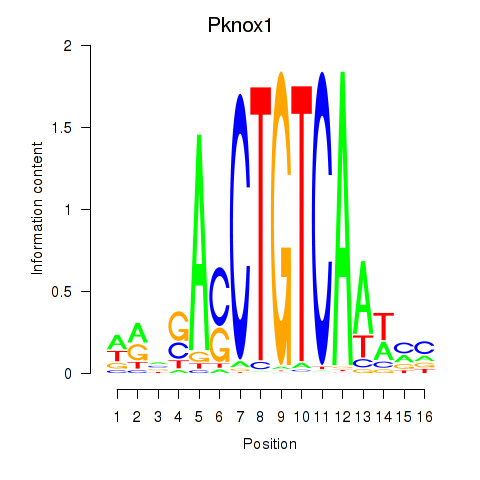
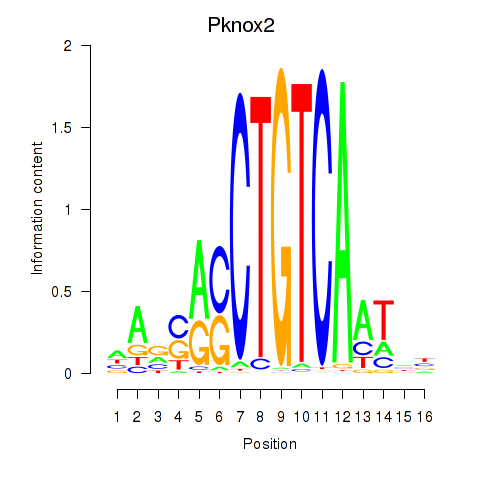
Transcription factors associated with Pknox2_Pknox1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Pknox1 | ENSMUSG00000006705.6 | Pknox1 |
| Pknox2 | ENSMUSG00000035934.9 | Pknox2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pknox1 | mm10_v2_chr17_+_31564749_31564854 | 0.67 | 6.7e-09 | Click! |
| Pknox2 | mm10_v2_chr9_-_37147257_37147323 | 0.06 | 6.8e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.4 | GO:1901228 | regulation of osteoclast proliferation(GO:0090289) negative regulation of bone mineralization involved in bone maturation(GO:1900158) positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 1.2 | 7.1 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 1.0 | 9.8 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.8 | 4.8 | GO:0035743 | CD4-positive, alpha-beta T cell cytokine production(GO:0035743) T-helper 2 cell cytokine production(GO:0035745) |
| 0.6 | 4.5 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.6 | 1.9 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.6 | 7.0 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.6 | 3.7 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.5 | 1.4 | GO:0030070 | insulin processing(GO:0030070) |
| 0.4 | 5.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.4 | 1.5 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.4 | 2.5 | GO:1905206 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) positive regulation of hydrogen peroxide-induced cell death(GO:1905206) |
| 0.3 | 2.3 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.3 | 1.6 | GO:0021905 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) |
| 0.3 | 2.3 | GO:0072539 | T-helper 17 cell differentiation(GO:0072539) |
| 0.3 | 0.8 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.2 | 1.7 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.2 | 0.8 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.2 | 1.5 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.2 | 0.9 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.2 | 0.7 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 1.0 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.1 | 1.3 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 1.5 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 0.3 | GO:2000813 | negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.1 | 0.9 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.1 | 0.4 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.1 | 1.2 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.1 | 0.6 | GO:2000325 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.1 | 2.9 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 0.7 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 0.7 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 | 1.5 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.1 | 1.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.2 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.1 | 5.4 | GO:0045666 | positive regulation of neuron differentiation(GO:0045666) |
| 0.0 | 1.1 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 1.0 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 3.4 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 4.6 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.5 | GO:0044144 | regulation of growth of symbiont in host(GO:0044126) modulation of growth of symbiont involved in interaction with host(GO:0044144) |
| 0.0 | 0.2 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 2.9 | GO:0045638 | negative regulation of myeloid cell differentiation(GO:0045638) |
| 0.0 | 2.0 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.7 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.9 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 1.3 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.9 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 1.1 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.5 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.1 | GO:2000252 | negative regulation of feeding behavior(GO:2000252) |
| 0.0 | 0.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 6.4 | GO:0006897 | endocytosis(GO:0006897) |
| 0.0 | 2.5 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.0 | 0.2 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 1.1 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.6 | 7.0 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.5 | 5.9 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.4 | 3.7 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.2 | 4.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 2.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 1.5 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 1.5 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.9 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 0.9 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 0.8 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.1 | 1.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 1.0 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 0.3 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 6.2 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.8 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.3 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.3 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.7 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 2.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.6 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 1.3 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 4.9 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.7 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 3.7 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 6.9 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 4.2 | GO:0009986 | cell surface(GO:0009986) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.1 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 1.6 | 9.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.9 | 5.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.6 | 2.3 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.5 | 7.0 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.5 | 4.4 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.3 | 1.0 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.2 | 2.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.2 | 1.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.2 | 6.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 1.6 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 1.5 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 3.7 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.1 | 0.8 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.1 | 4.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.4 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) cobalt ion binding(GO:0050897) |
| 0.1 | 1.0 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.9 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.1 | 0.7 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 5.4 | GO:0020037 | heme binding(GO:0020037) |
| 0.1 | 0.7 | GO:0052872 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) phenanthrene 9,10-monooxygenase activity(GO:0018636) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) tocotrienol omega-hydroxylase activity(GO:0052872) thalianol hydroxylase activity(GO:0080014) |
| 0.1 | 1.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 1.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 1.6 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 0.2 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.1 | 2.5 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.1 | 4.5 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 2.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.7 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 1.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 4.8 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 2.5 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 1.9 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 1.5 | GO:0032934 | cholesterol binding(GO:0015485) sterol binding(GO:0032934) |
| 0.0 | 0.4 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.5 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.8 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 5.2 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.9 | GO:0008026 | ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.0 | 0.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 1.3 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 0.4 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.0 | 1.3 | GO:0043851 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) rRNA (uridine-2'-O-)-methyltransferase activity(GO:0008650) rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) selenocysteine methyltransferase activity(GO:0016205) rRNA (adenine) methyltransferase activity(GO:0016433) rRNA (cytosine) methyltransferase activity(GO:0016434) rRNA (guanine) methyltransferase activity(GO:0016435) 1-phenanthrol methyltransferase activity(GO:0018707) protein-arginine N5-methyltransferase activity(GO:0019702) dimethylarsinite methyltransferase activity(GO:0034541) 4,5-dihydroxybenzo(a)pyrene methyltransferase activity(GO:0034807) 1-hydroxypyrene methyltransferase activity(GO:0034931) 1-hydroxy-6-methoxypyrene methyltransferase activity(GO:0034933) demethylmenaquinone methyltransferase activity(GO:0043770) cobalt-precorrin-6B C5-methyltransferase activity(GO:0043776) cobalt-precorrin-7 C15-methyltransferase activity(GO:0043777) cobalt-precorrin-5B C1-methyltransferase activity(GO:0043780) cobalt-precorrin-3 C17-methyltransferase activity(GO:0043782) dimethylamine methyltransferase activity(GO:0043791) hydroxyneurosporene-O-methyltransferase activity(GO:0043803) tRNA (adenine-57, 58-N(1)-) methyltransferase activity(GO:0043827) methylamine-specific methylcobalamin:coenzyme M methyltransferase activity(GO:0043833) trimethylamine methyltransferase activity(GO:0043834) methanol-specific methylcobalamin:coenzyme M methyltransferase activity(GO:0043851) monomethylamine methyltransferase activity(GO:0043852) P-methyltransferase activity(GO:0051994) Se-methyltransferase activity(GO:0051995) 2-phytyl-1,4-naphthoquinone methyltransferase activity(GO:0052624) tRNA (uracil-2'-O-)-methyltransferase activity(GO:0052665) tRNA (cytosine-2'-O-)-methyltransferase activity(GO:0052666) phosphomethylethanolamine N-methyltransferase activity(GO:0052667) tRNA (cytosine-3-)-methyltransferase activity(GO:0052735) rRNA (cytosine-2'-O-)-methyltransferase activity(GO:0070677) rRNA (cytosine-N4-)-methyltransferase activity(GO:0071424) trihydroxyferuloyl spermidine O-methyltransferase activity(GO:0080012) |
| 0.0 | 9.8 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) |
| 0.0 | 0.2 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |