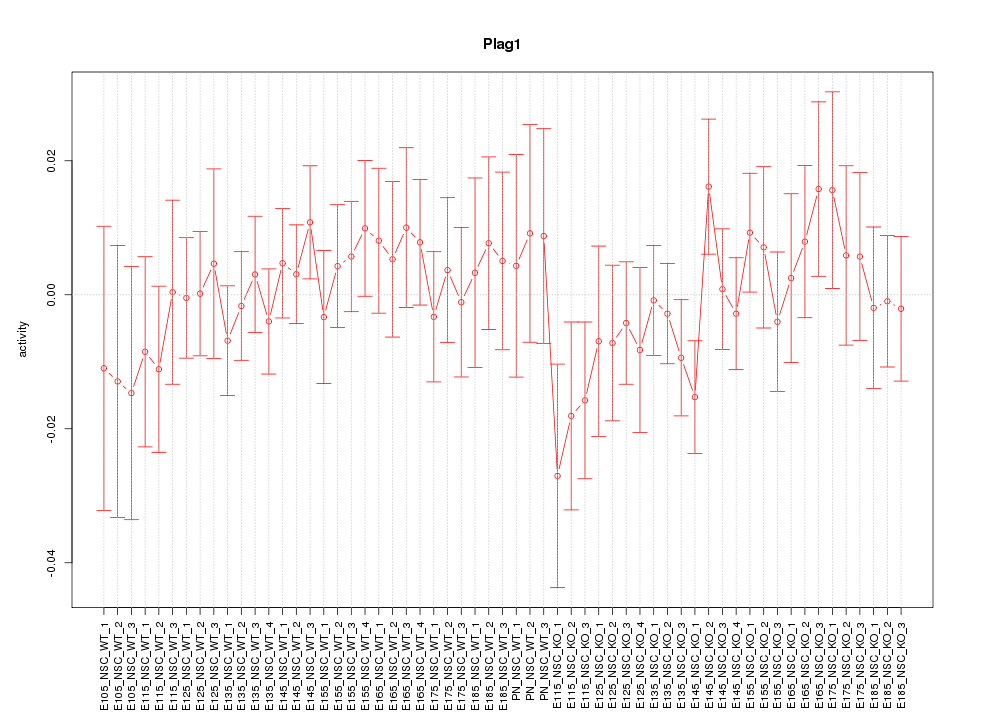
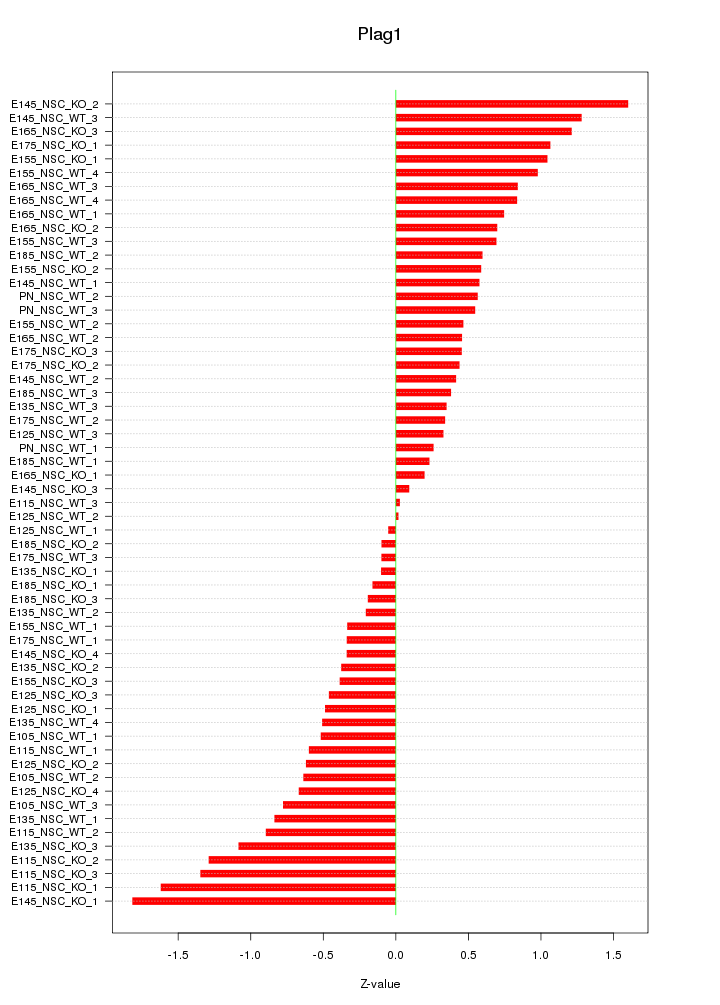
Motif ID: Plag1
Z-value: 0.726
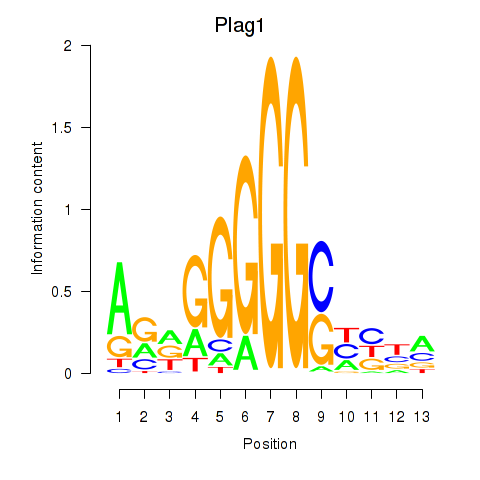
Transcription factors associated with Plag1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Plag1 | ENSMUSG00000003282.3 | Plag1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Plag1 | mm10_v2_chr4_-_3938354_3938401 | -0.70 | 8.8e-10 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 13.6 | GO:0030827 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 2.0 | 8.2 | GO:1901204 | positive regulation of guanylate cyclase activity(GO:0031284) regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 1.3 | 3.8 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 1.1 | 7.7 | GO:0032811 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) negative regulation of epinephrine secretion(GO:0032811) |
| 1.1 | 5.4 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.9 | 5.6 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.8 | 4.0 | GO:0061438 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) |
| 0.7 | 3.0 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.7 | 2.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.7 | 2.7 | GO:0003357 | noradrenergic neuron differentiation(GO:0003357) |
| 0.6 | 1.7 | GO:0070671 | response to interleukin-12(GO:0070671) |
| 0.6 | 1.7 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.6 | 2.3 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.5 | 2.1 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.5 | 8.0 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.5 | 1.6 | GO:0032513 | negative regulation of protein phosphatase type 2B activity(GO:0032513) |
| 0.5 | 3.6 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.5 | 0.9 | GO:0021933 | radial glia guided migration of cerebellar granule cell(GO:0021933) |
| 0.5 | 0.5 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.5 | 2.3 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.4 | 2.2 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.4 | 1.8 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.4 | 1.2 | GO:0060084 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) circadian sleep/wake cycle, REM sleep(GO:0042747) synaptic transmission involved in micturition(GO:0060084) |
| 0.4 | 10.2 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.4 | 1.2 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.4 | 1.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.4 | 3.5 | GO:0090494 | catecholamine uptake(GO:0090493) dopamine uptake(GO:0090494) |
| 0.4 | 1.2 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.4 | 2.3 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.4 | 0.8 | GO:0010958 | regulation of amino acid import(GO:0010958) regulation of L-arginine import(GO:0010963) |
| 0.4 | 4.2 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.4 | 1.1 | GO:1901254 | positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.4 | 2.2 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.3 | 1.0 | GO:1903903 | regulation of establishment of T cell polarity(GO:1903903) |
| 0.3 | 1.0 | GO:0003195 | tricuspid valve development(GO:0003175) tricuspid valve morphogenesis(GO:0003186) tricuspid valve formation(GO:0003195) right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.3 | 1.0 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.3 | 1.6 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.3 | 3.2 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.3 | 2.2 | GO:1903423 | positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.3 | 1.2 | GO:1904529 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.3 | 1.2 | GO:0071692 | protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) |
| 0.3 | 0.3 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.3 | 1.5 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.3 | 1.5 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.3 | 0.9 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.3 | 1.9 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.3 | 0.8 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.3 | 0.8 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.3 | 1.5 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.2 | 1.7 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.2 | 2.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.2 | 12.3 | GO:0021696 | cerebellar cortex morphogenesis(GO:0021696) |
| 0.2 | 0.7 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.2 | 2.6 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.2 | 2.7 | GO:0097688 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.2 | 0.9 | GO:0061428 | positive regulation of oocyte development(GO:0060282) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.2 | 1.5 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.2 | 2.4 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.2 | 4.7 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.2 | 0.6 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.2 | 0.6 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.2 | 0.6 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.2 | 1.0 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.2 | 4.0 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.2 | 1.0 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.2 | 0.8 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.2 | 1.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.2 | 1.2 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.2 | 0.8 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) |
| 0.2 | 1.1 | GO:0071415 | cellular response to caffeine(GO:0071313) cellular response to purine-containing compound(GO:0071415) |
| 0.2 | 1.3 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.2 | 1.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.2 | 4.9 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.2 | 1.7 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.2 | 1.7 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.2 | 0.6 | GO:0048611 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.2 | 0.6 | GO:0043181 | vacuolar sequestering(GO:0043181) |
| 0.2 | 1.7 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.2 | 0.7 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.2 | 0.9 | GO:0045188 | optic nerve morphogenesis(GO:0021631) regulation of circadian sleep/wake cycle, non-REM sleep(GO:0045188) |
| 0.2 | 0.7 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.2 | 1.8 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.2 | 1.0 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.2 | 1.0 | GO:1902998 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of tau-protein kinase activity(GO:1902949) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.2 | 0.2 | GO:2000277 | positive regulation of oxidative phosphorylation uncoupler activity(GO:2000277) |
| 0.2 | 0.8 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.2 | 0.5 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.2 | 0.5 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.2 | 1.0 | GO:1903056 | regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.2 | 1.0 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 1.9 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.2 | 1.4 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.2 | 0.8 | GO:0043314 | negative regulation of cellular extravasation(GO:0002692) negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.2 | 0.8 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.2 | 1.6 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.2 | 0.2 | GO:0002578 | negative regulation of antigen processing and presentation(GO:0002578) |
| 0.2 | 0.8 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.2 | 0.5 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.2 | 0.5 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 0.7 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.1 | 1.5 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.1 | 0.9 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.1 | 0.4 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 2.9 | GO:0095500 | acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 | 0.4 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.1 | 0.6 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 0.7 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.1 | 0.4 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.1 | 2.5 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.1 | 1.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.8 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 0.4 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 | 1.7 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) |
| 0.1 | 0.4 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.1 | 0.7 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.1 | 3.8 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.1 | 1.0 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 0.8 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 1.4 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.9 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.1 | 1.2 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.1 | 1.5 | GO:0015816 | glycine transport(GO:0015816) |
| 0.1 | 1.6 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 0.6 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 0.4 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 | 0.6 | GO:0021636 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) |
| 0.1 | 0.5 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 0.5 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.1 | 1.1 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.1 | 0.5 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 4.4 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.1 | 0.5 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.1 | 0.2 | GO:0010533 | regulation of activation of Janus kinase activity(GO:0010533) regulation of activation of JAK2 kinase activity(GO:0010534) |
| 0.1 | 0.8 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 0.6 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.1 | 0.8 | GO:0052490 | suppression by virus of host apoptotic process(GO:0019050) negative regulation by symbiont of host apoptotic process(GO:0033668) modulation by virus of host apoptotic process(GO:0039526) negative regulation by symbiont of host programmed cell death(GO:0052041) negative regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052490) |
| 0.1 | 0.5 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.1 | 0.7 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 1.2 | GO:0001956 | positive regulation of neurotransmitter secretion(GO:0001956) positive regulation of neurotransmitter transport(GO:0051590) |
| 0.1 | 0.3 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.1 | 2.7 | GO:0010043 | response to zinc ion(GO:0010043) |
| 0.1 | 1.0 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 0.5 | GO:0044144 | regulation of growth of symbiont in host(GO:0044126) modulation of growth of symbiont involved in interaction with host(GO:0044144) |
| 0.1 | 0.8 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 | 0.4 | GO:0006842 | tricarboxylic acid transport(GO:0006842) succinate transport(GO:0015744) citrate transport(GO:0015746) |
| 0.1 | 0.3 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.1 | 1.0 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 1.4 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.1 | 0.6 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.3 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.1 | 0.4 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.1 | 1.1 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 0.7 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.1 | 0.7 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.1 | 1.5 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 0.3 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.1 | 2.0 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.1 | 0.4 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.1 | 0.6 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.1 | 0.4 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.6 | GO:2000505 | regulation of energy homeostasis(GO:2000505) |
| 0.1 | 1.6 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 | 0.8 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 8.4 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 0.9 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.1 | 0.4 | GO:0043388 | positive regulation of DNA binding(GO:0043388) |
| 0.1 | 0.4 | GO:1904417 | regulation of xenophagy(GO:1904415) positive regulation of xenophagy(GO:1904417) |
| 0.1 | 0.9 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 1.1 | GO:0072010 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.1 | 2.6 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 1.1 | GO:0009072 | aromatic amino acid family metabolic process(GO:0009072) |
| 0.1 | 3.8 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 | 0.6 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.5 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 | 1.0 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.1 | 0.7 | GO:1901030 | positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901030) |
| 0.1 | 0.5 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 0.1 | 0.3 | GO:0021886 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.1 | 0.4 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.1 | 1.0 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.1 | 0.3 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 0.9 | GO:0046321 | positive regulation of fatty acid oxidation(GO:0046321) |
| 0.1 | 1.0 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.1 | 0.3 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 0.5 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.1 | 0.2 | GO:0070445 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.1 | 0.2 | GO:2000501 | natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.1 | 4.1 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.1 | 1.7 | GO:2001258 | negative regulation of cation channel activity(GO:2001258) |
| 0.1 | 1.6 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.1 | 0.9 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.1 | 0.2 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.1 | 0.5 | GO:0071225 | cellular response to muramyl dipeptide(GO:0071225) |
| 0.1 | 1.0 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.1 | 0.4 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.7 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.1 | 0.3 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.1 | 0.7 | GO:0035646 | endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.1 | 0.6 | GO:1904587 | glycoprotein ERAD pathway(GO:0097466) response to glycoprotein(GO:1904587) |
| 0.1 | 0.4 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.1 | 1.9 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 1.1 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 0.5 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.1 | 0.2 | GO:0033092 | positive regulation of immature T cell proliferation(GO:0033091) positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.1 | 0.3 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.1 | 1.1 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 0.7 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.1 | 0.3 | GO:0043206 | extracellular fibril organization(GO:0043206) |
| 0.1 | 0.9 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.2 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.1 | 0.4 | GO:0003413 | chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) |
| 0.1 | 0.2 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.1 | 0.6 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.1 | 0.3 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.1 | 0.2 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.1 | 1.8 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 0.3 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.1 | 0.6 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.6 | GO:0060326 | cell chemotaxis(GO:0060326) |
| 0.0 | 0.0 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 0.4 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.7 | GO:1900273 | positive regulation of long-term synaptic potentiation(GO:1900273) |
| 0.0 | 1.4 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.6 | GO:0043496 | regulation of protein homodimerization activity(GO:0043496) |
| 0.0 | 0.9 | GO:0071357 | type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.4 | GO:0014898 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 | 0.5 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) regulation of histone H3-K9 trimethylation(GO:1900112) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 1.2 | GO:0015991 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 0.3 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 1.1 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.8 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.1 | GO:0097274 | urea homeostasis(GO:0097274) |
| 0.0 | 0.6 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.1 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 1.4 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 0.7 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.2 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 1.0 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.1 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 0.0 | 0.3 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.2 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.0 | 0.1 | GO:0090148 | membrane fission(GO:0090148) |
| 0.0 | 0.0 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.0 | 0.1 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.0 | 1.1 | GO:0045453 | bone resorption(GO:0045453) |
| 0.0 | 0.3 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.5 | GO:0032148 | activation of protein kinase B activity(GO:0032148) |
| 0.0 | 1.6 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.5 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.0 | 0.8 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.3 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.0 | 1.3 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.0 | 0.1 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.0 | 0.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.1 | GO:0002182 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.0 | 0.2 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.0 | 0.2 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.5 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 2.0 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.0 | 0.5 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.2 | GO:0036230 | granulocyte activation(GO:0036230) |
| 0.0 | 0.1 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.1 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.0 | 0.1 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.4 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.2 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.2 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.7 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.1 | GO:0019249 | lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) |
| 0.0 | 0.5 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.0 | 0.3 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.2 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.8 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.2 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.0 | 0.1 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.0 | 0.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 2.4 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 1.1 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.0 | 0.1 | GO:0071028 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 0.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.4 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.1 | GO:1900194 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.8 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.2 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.0 | 0.2 | GO:0032095 | regulation of response to food(GO:0032095) |
| 0.0 | 0.2 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.7 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.6 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.4 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.1 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.2 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.7 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 | 0.2 | GO:0034088 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.0 | 0.1 | GO:0072362 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) regulation of lipid transport by negative regulation of transcription from RNA polymerase II promoter(GO:0072368) |
| 0.0 | 0.1 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.3 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 0.1 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 0.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.3 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.4 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.1 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.0 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) negative regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902548) |
| 0.0 | 0.4 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.2 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.5 | GO:0033014 | porphyrin-containing compound biosynthetic process(GO:0006779) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.0 | 0.4 | GO:0019915 | lipid storage(GO:0019915) |
| 0.0 | 0.4 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.0 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 0.2 | GO:0042491 | auditory receptor cell differentiation(GO:0042491) |
| 0.0 | 0.3 | GO:0051220 | cytoplasmic sequestering of protein(GO:0051220) |
| 0.0 | 0.1 | GO:0032091 | negative regulation of protein binding(GO:0032091) |
| 0.0 | 0.3 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 7.7 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 1.5 | 14.7 | GO:0044327 | dendritic spine head(GO:0044327) |
| 1.1 | 3.4 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 1.1 | 6.7 | GO:0008091 | spectrin(GO:0008091) |
| 0.8 | 2.3 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.7 | 2.0 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.7 | 2.0 | GO:0005595 | collagen type XII trimer(GO:0005595) anchoring collagen complex(GO:0030934) |
| 0.6 | 1.9 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.6 | 6.8 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.5 | 1.4 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.4 | 1.6 | GO:1902737 | dendritic filopodium(GO:1902737) |
| 0.4 | 5.4 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.3 | 5.2 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.3 | 4.3 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.3 | 1.5 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.3 | 4.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 1.0 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.2 | 1.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 2.1 | GO:0042581 | specific granule(GO:0042581) |
| 0.2 | 1.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.2 | 1.3 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.2 | 0.9 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.2 | 0.4 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.2 | 1.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.2 | 1.0 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.2 | 3.9 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.2 | 2.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.2 | 9.6 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.2 | 1.3 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.2 | 0.5 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.2 | 1.0 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) neurofibrillary tangle(GO:0097418) |
| 0.2 | 2.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.2 | 2.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 4.7 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.2 | 0.5 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 1.5 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 3.8 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.1 | 1.4 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 0.3 | GO:0044299 | C-fiber(GO:0044299) |
| 0.1 | 4.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 0.7 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 2.3 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 1.0 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 1.9 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 1.4 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 1.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.8 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 0.6 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 0.6 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 1.6 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 1.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 1.2 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.1 | 1.2 | GO:0001527 | microfibril(GO:0001527) |
| 0.1 | 1.0 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 0.7 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 4.8 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.1 | 0.4 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 0.3 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 3.4 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 0.7 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 0.7 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.3 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.1 | 1.0 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 0.3 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.1 | 0.8 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 2.5 | GO:0044853 | plasma membrane raft(GO:0044853) |
| 0.1 | 0.5 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 1.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 0.2 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 0.1 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.1 | 0.8 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 0.2 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 1.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 1.3 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 1.3 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 1.6 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.3 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 0.5 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.1 | 0.3 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 2.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.9 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.1 | 0.5 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.1 | 1.0 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.1 | 0.6 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 1.9 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.5 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.6 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.9 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 2.6 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 1.5 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.1 | GO:0098576 | lumenal side of membrane(GO:0098576) |
| 0.0 | 0.6 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.0 | 10.9 | GO:0097060 | synaptic membrane(GO:0097060) |
| 0.0 | 4.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.4 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.4 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.6 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.6 | GO:0032420 | stereocilium(GO:0032420) actin-based cell projection(GO:0098858) |
| 0.0 | 0.4 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 1.1 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 1.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 3.1 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.7 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.2 | GO:0071010 | prespliceosome(GO:0071010) |
| 0.0 | 1.1 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.8 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.2 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 3.5 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.0 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 4.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.3 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.1 | GO:0090537 | CERF complex(GO:0090537) |
| 0.0 | 2.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.7 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.4 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 1.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.0 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.4 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.7 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.9 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.4 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.2 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.0 | 0.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.8 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.0 | 0.1 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.0 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.8 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 1.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.6 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 0.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 6.5 | GO:0044297 | cell body(GO:0044297) |
| 0.0 | 3.0 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 1.6 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 5.1 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.1 | GO:0043195 | terminal bouton(GO:0043195) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.2 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 1.4 | 4.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 1.3 | 7.7 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.9 | 2.7 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.9 | 3.6 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.8 | 4.0 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.8 | 3.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.7 | 5.2 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.6 | 5.4 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.6 | 2.3 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.5 | 2.3 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.4 | 0.9 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.4 | 2.2 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.4 | 2.5 | GO:0019841 | retinol binding(GO:0019841) |
| 0.4 | 3.7 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.4 | 3.6 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.4 | 1.6 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.4 | 3.9 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.4 | 1.6 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.4 | 1.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.4 | 6.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.4 | 2.9 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.4 | 1.1 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.4 | 1.8 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.3 | 1.0 | GO:0036361 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.3 | 1.0 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.3 | 2.7 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.3 | 1.0 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.3 | 1.0 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
| 0.3 | 0.6 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.3 | 3.5 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.3 | 1.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.3 | 2.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.3 | 0.8 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.3 | 0.8 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.3 | 1.8 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.3 | 1.0 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.2 | 1.5 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.2 | 1.0 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.2 | 0.9 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.2 | 1.9 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.2 | 2.3 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.2 | 3.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 0.9 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.2 | 2.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 5.0 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.2 | 4.1 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.2 | 3.2 | GO:0008430 | selenium binding(GO:0008430) |
| 0.2 | 0.6 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.2 | 0.8 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 1.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.2 | 0.6 | GO:0032450 | oligo-1,6-glucosidase activity(GO:0004574) maltose alpha-glucosidase activity(GO:0032450) |
| 0.2 | 0.4 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.2 | 2.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.2 | 1.8 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.2 | 0.9 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.2 | 3.0 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.2 | 1.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.2 | 0.7 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.2 | 3.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.2 | 1.0 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.2 | 0.5 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.2 | 0.5 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.1 | 0.6 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.9 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.1 | 1.0 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.1 | 3.8 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 1.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.8 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.4 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.1 | 1.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.8 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 0.6 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 0.5 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.1 | 5.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 1.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 1.2 | GO:0004889 | acetylcholine-activated cation-selective channel activity(GO:0004889) acetylcholine binding(GO:0042166) |
| 0.1 | 0.6 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 1.2 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.1 | 0.7 | GO:0046978 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.1 | 0.6 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.6 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 2.7 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 1.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 1.1 | GO:0034943 | acyl-CoA ligase activity(GO:0003996) 3-oxo-2-(2'-pentenyl)cyclopentane-1-octanoic acid CoA ligase activity(GO:0010435) 3-isopropenyl-6-oxoheptanoyl-CoA synthetase activity(GO:0018854) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA synthetase activity(GO:0018855) benzoyl acetate-CoA ligase activity(GO:0018856) 2,4-dichlorobenzoate-CoA ligase activity(GO:0018857) very long-chain fatty acid-CoA ligase activity(GO:0031957) pivalate-CoA ligase activity(GO:0034783) cyclopropanecarboxylate-CoA ligase activity(GO:0034793) adipate-CoA ligase activity(GO:0034796) citronellyl-CoA ligase activity(GO:0034823) mentha-1,3-dione-CoA ligase activity(GO:0034841) thiophene-2-carboxylate-CoA ligase activity(GO:0034842) 2,4,4-trimethylpentanoate-CoA ligase activity(GO:0034865) cis-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034942) trans-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034943) branched-chain acyl-CoA synthetase (ADP-forming) activity(GO:0043759) aryl-CoA synthetase (ADP-forming) activity(GO:0043762) 3-hydroxypropionyl-CoA synthetase activity(GO:0043955) perillic acid:CoA ligase (ADP-forming) activity(GO:0052685) perillic acid:CoA ligase (AMP-forming) activity(GO:0052686) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (ADP-forming) activity(GO:0052687) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (AMP-forming) activity(GO:0052688) pristanate-CoA ligase activity(GO:0070251) malonyl-CoA synthetase activity(GO:0090409) |
| 0.1 | 0.2 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.1 | 2.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 1.4 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.6 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.1 | 1.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.3 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 0.6 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.1 | 3.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.4 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.1 | 0.6 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 0.5 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 1.0 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 3.4 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 0.8 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.1 | 0.4 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.1 | 0.5 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 0.8 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 0.9 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.8 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.1 | 0.5 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.1 | 3.7 | GO:0005267 | potassium channel activity(GO:0005267) |
| 0.1 | 0.3 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.1 | 2.8 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.4 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.1 | 0.8 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.1 | 2.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.8 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 1.7 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 0.3 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.4 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.2 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.1 | 1.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 0.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.3 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.1 | 0.3 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 0.5 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 0.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.1 | 2.0 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 1.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 1.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 0.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 0.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.2 | GO:0070330 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) aromatase activity(GO:0070330) |
| 0.1 | 0.2 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 0.3 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 2.0 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.3 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.6 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 0.2 | GO:0008521 | acetyl-CoA transporter activity(GO:0008521) |
| 0.1 | 0.4 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.2 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.9 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 1.1 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.3 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 1.0 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.4 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.5 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.9 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.2 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.0 | 0.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 1.2 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 1.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.6 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.8 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.9 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.4 | GO:0018602 | sulfonate dioxygenase activity(GO:0000907) 2,4-dichlorophenoxyacetate alpha-ketoglutarate dioxygenase activity(GO:0018602) hypophosphite dioxygenase activity(GO:0034792) gibberellin 2-beta-dioxygenase activity(GO:0045543) C-19 gibberellin 2-beta-dioxygenase activity(GO:0052634) C-20 gibberellin 2-beta-dioxygenase activity(GO:0052635) |
| 0.0 | 0.5 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:0019002 | GMP binding(GO:0019002) |
| 0.0 | 0.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.7 | GO:0035380 | C-3 sterol dehydrogenase (C-4 sterol decarboxylase) activity(GO:0000252) mevaldate reductase activity(GO:0004495) gluconate dehydrogenase activity(GO:0008875) epoxide dehydrogenase activity(GO:0018451) 5-exo-hydroxycamphor dehydrogenase activity(GO:0018452) 2-hydroxytetrahydrofuran dehydrogenase activity(GO:0018453) acetoin dehydrogenase activity(GO:0019152) phenylcoumaran benzylic ether reductase activity(GO:0032442) D-xylose:NADP reductase activity(GO:0032866) L-arabinose:NADP reductase activity(GO:0032867) D-arabinitol dehydrogenase, D-ribulose forming (NADP+) activity(GO:0033709) (R)-(-)-1,2,3,4-tetrahydronaphthol dehydrogenase activity(GO:0034831) 3-hydroxymenthone dehydrogenase activity(GO:0034840) very long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0035380) dihydrotestosterone 17-beta-dehydrogenase activity(GO:0035410) (R)-2-hydroxyisocaproate dehydrogenase activity(GO:0043713) L-arabinose 1-dehydrogenase (NADP+) activity(GO:0044103) L-xylulose reductase (NAD+) activity(GO:0044105) 3-ketoglucose-reductase activity(GO:0048258) D-arabinitol dehydrogenase, D-xylulose forming (NADP+) activity(GO:0052677) |
| 0.0 | 0.3 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 1.3 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 1.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.3 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.2 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.1 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.8 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.2 | GO:0045703 | pinocarveol dehydrogenase activity(GO:0018446) chloral hydrate dehydrogenase activity(GO:0018447) hydroxymethylmethylsilanediol oxidase activity(GO:0018448) 1-phenylethanol dehydrogenase activity(GO:0018449) myrtenol dehydrogenase activity(GO:0018450) cis-1,2-dihydroxy-1,2-dihydro-8-carboxynaphthalene dehydrogenase activity(GO:0034522) 3-hydroxy-4-methyloctanoyl-CoA dehydrogenase activity(GO:0034582) 2-hydroxy-4-isopropenylcyclohexane-1-carboxyl-CoA dehydrogenase activity(GO:0034778) cis-9,10-dihydroanthracene-9,10-diol dehydrogenase activity(GO:0034817) citronellol dehydrogenase activity(GO:0034821) naphthyl-2-hydroxymethyl-succinyl-CoA dehydrogenase activity(GO:0034847) 2,4,4-trimethyl-1-pentanol dehydrogenase activity(GO:0034863) 2,4,4-trimethyl-3-hydroxypentanoyl-CoA dehydrogenase activity(GO:0034868) 1-hydroxy-4,4-dimethylpentan-3-one dehydrogenase activity(GO:0034871) endosulfan diol dehydrogenase activity(GO:0034891) endosulfan hydroxyether dehydrogenase activity(GO:0034901) 3-hydroxy-2-methylhexanoyl-CoA dehydrogenase activity(GO:0034918) 3-hydroxy-2,6-dimethyl-5-methylene-heptanoyl-CoA dehydrogenase activity(GO:0034944) versicolorin reductase activity(GO:0042469) ketoreductase activity(GO:0045703) |
| 0.0 | 0.2 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.6 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.0 | 0.2 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.3 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.1 | GO:0070615 | nucleosome-dependent ATPase activity(GO:0070615) |
| 0.0 | 0.4 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.4 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.4 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.1 | GO:0086038 | calcium:sodium antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086038) |
| 0.0 | 0.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.2 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.2 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.2 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.7 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.5 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.1 | GO:0050827 | toxin receptor binding(GO:0050827) |
| 0.0 | 1.7 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) |
| 0.0 | 0.3 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 1.4 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 5.7 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.5 | GO:0005230 | extracellular ligand-gated ion channel activity(GO:0005230) |
| 0.0 | 0.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 1.4 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.7 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.0 | 1.4 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 0.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.4 | GO:0052771 | coenzyme F390-A hydrolase activity(GO:0052770) coenzyme F390-G hydrolase activity(GO:0052771) |