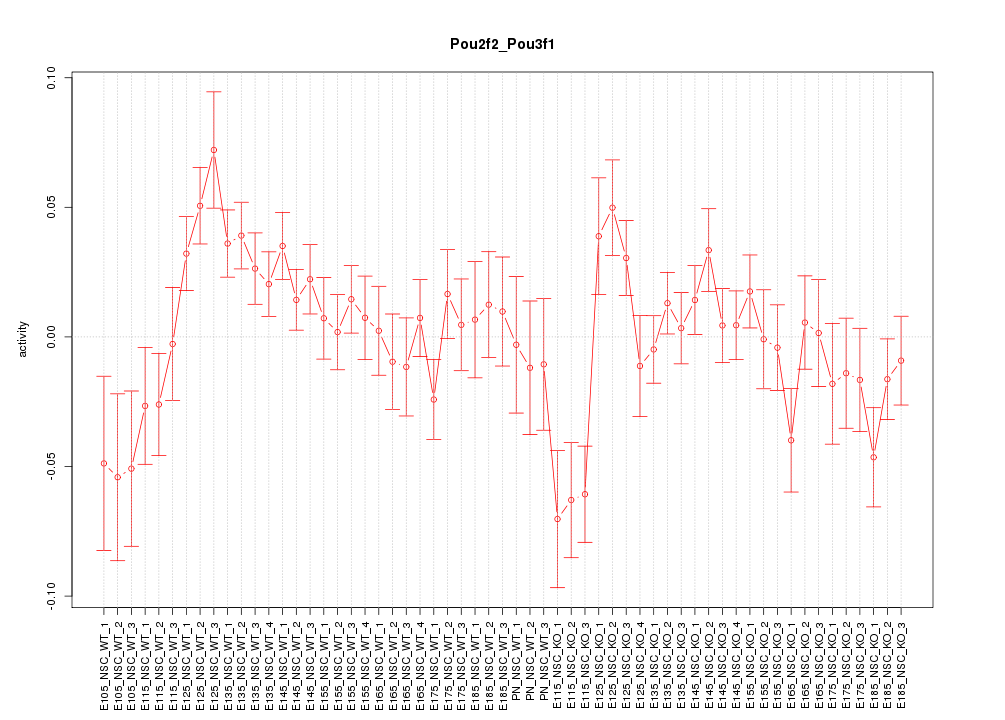
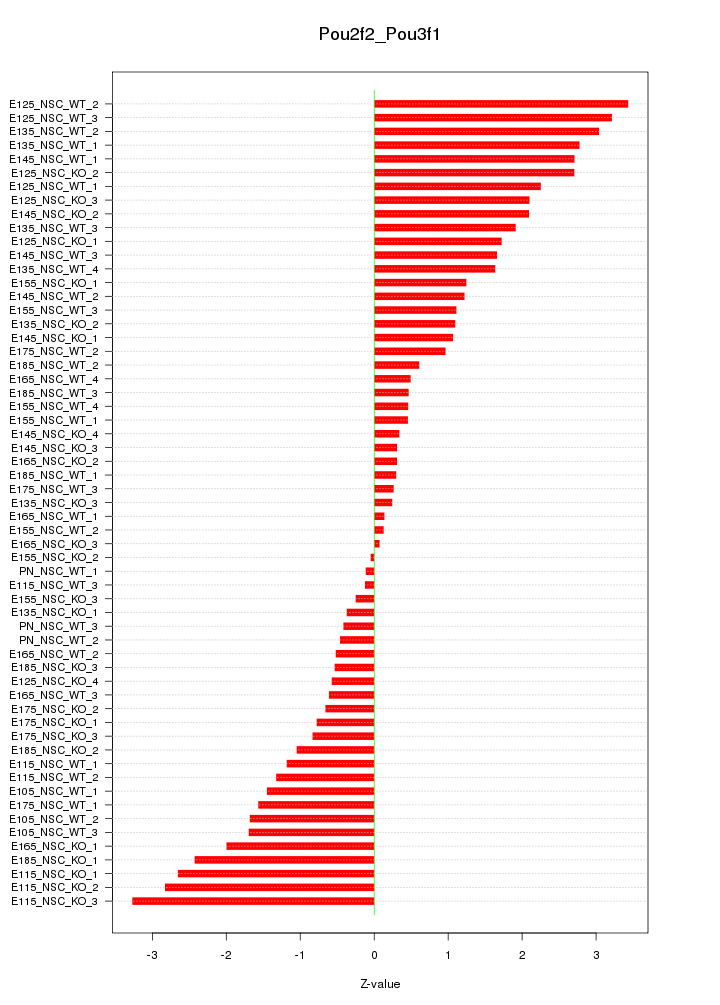
Motif ID: Pou2f2_Pou3f1
Z-value: 1.556
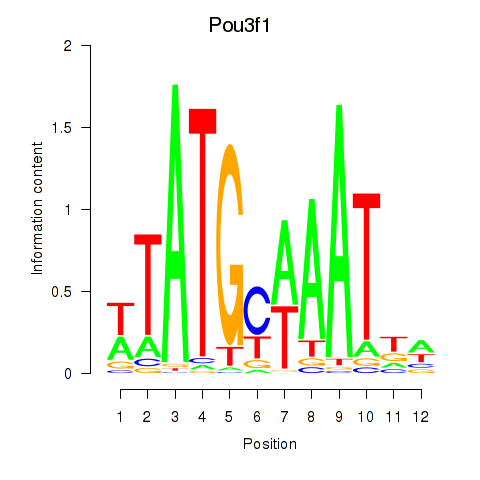
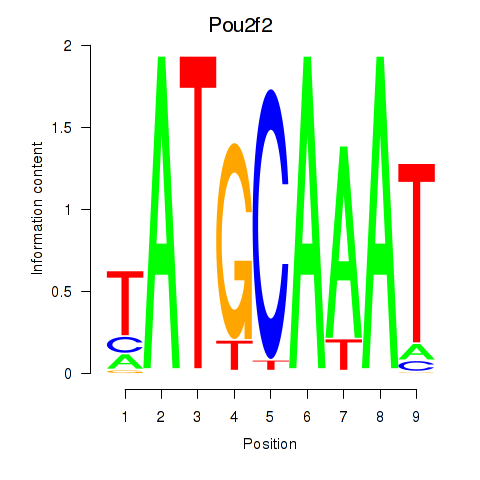
Transcription factors associated with Pou2f2_Pou3f1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Pou2f2 | ENSMUSG00000008496.12 | Pou2f2 |
| Pou3f1 | ENSMUSG00000090125.2 | Pou3f1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou3f1 | mm10_v2_chr4_+_124657646_124657656 | 0.28 | 3.3e-02 | Click! |
| Pou2f2 | mm10_v2_chr7_-_25132473_25132512 | 0.13 | 3.4e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 8.4 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 2.1 | 10.4 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 1.7 | 185.2 | GO:0006342 | chromatin silencing(GO:0006342) |
| 1.7 | 20.1 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 1.5 | 7.7 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 1.2 | 3.5 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.9 | 2.7 | GO:0021893 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 0.9 | 6.1 | GO:0034047 | regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.8 | 2.5 | GO:0045014 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.7 | 4.7 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.7 | 4.0 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.6 | 2.9 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.6 | 1.7 | GO:0060853 | arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) endothelial cell fate specification(GO:0060847) Notch signaling pathway involved in arterial endothelial cell fate commitment(GO:0060853) blood vessel endothelial cell fate specification(GO:0097101) |
| 0.5 | 5.5 | GO:0042095 | interferon-gamma biosynthetic process(GO:0042095) |
| 0.5 | 3.5 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.5 | 3.0 | GO:0035743 | CD4-positive, alpha-beta T cell cytokine production(GO:0035743) T-helper 2 cell cytokine production(GO:0035745) |
| 0.4 | 1.8 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.4 | 3.3 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.4 | 11.6 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.3 | 1.4 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.3 | 11.5 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.3 | 5.5 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.3 | 1.3 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.3 | 2.5 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.3 | 26.1 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.3 | 1.8 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.2 | 1.3 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.2 | 4.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.2 | 3.5 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.2 | 0.8 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.2 | 1.1 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.2 | 0.5 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.2 | 1.3 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.2 | 0.7 | GO:2000110 | protein sialylation(GO:1990743) negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.2 | 1.4 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.2 | 6.0 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.2 | 1.5 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 0.4 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.1 | 2.3 | GO:0060065 | uterus development(GO:0060065) |
| 0.1 | 1.0 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 1.9 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.1 | 3.0 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 1.7 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 1.3 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.1 | 6.2 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.1 | 0.5 | GO:0007379 | segment specification(GO:0007379) |
| 0.1 | 0.5 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 0.4 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.1 | 0.2 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 0.0 | 0.8 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 1.7 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 1.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 1.0 | GO:0030262 | cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.4 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.2 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 3.0 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 4.2 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 0.2 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 2.9 | GO:0007612 | learning(GO:0007612) |
| 0.0 | 0.1 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.8 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.3 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 1.8 | GO:0048469 | cell maturation(GO:0048469) |
| 0.0 | 1.9 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.0 | 0.1 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 61.9 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 1.4 | 5.5 | GO:0002111 | BRCA2-BRAF35 complex(GO:0002111) |
| 1.1 | 45.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.9 | 8.9 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.9 | 3.5 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.6 | 4.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.3 | 127.7 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.3 | 6.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.3 | 1.4 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.2 | 2.9 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 4.2 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 0.8 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 7.9 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 5.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.8 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 5.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 0.7 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 1.3 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 4.1 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 2.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.4 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 2.6 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.8 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 3.0 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 1.1 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.5 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.8 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 8.4 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 1.0 | 4.0 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.8 | 4.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.7 | 4.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.4 | 6.1 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.4 | 1.5 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.3 | 2.5 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.3 | 1.4 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.2 | 2.9 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.2 | 1.7 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.2 | 1.3 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.2 | 4.3 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.2 | 7.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.2 | 1.8 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 2.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.2 | 1.9 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 1.0 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.7 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 1.8 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 1.0 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 3.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 67.3 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.1 | 10.2 | GO:0005496 | steroid binding(GO:0005496) |
| 0.1 | 1.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 0.4 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.1 | 3.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 0.8 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 6.1 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 8.8 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.1 | 0.4 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 1.2 | GO:0043747 | protein-N-terminal asparagine amidohydrolase activity(GO:0008418) UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase activity(GO:0008759) iprodione amidohydrolase activity(GO:0018748) (3,5-dichlorophenylurea)acetate amidohydrolase activity(GO:0018749) 4'-(2-hydroxyisopropyl)phenylurea amidohydrolase activity(GO:0034571) didemethylisoproturon amidohydrolase activity(GO:0034573) N-isopropylacetanilide amidohydrolase activity(GO:0034576) N-cyclohexylformamide amidohydrolase activity(GO:0034781) isonicotinic acid hydrazide hydrolase activity(GO:0034876) cis-aconitamide amidase activity(GO:0034882) gamma-N-formylaminovinylacetate hydrolase activity(GO:0034885) N2-acetyl-L-lysine deacetylase activity(GO:0043747) O-succinylbenzoate synthase activity(GO:0043748) indoleacetamide hydrolase activity(GO:0043864) N-acetylcitrulline deacetylase activity(GO:0043909) N-acetylgalactosamine-6-phosphate deacetylase activity(GO:0047419) diacetylchitobiose deacetylase activity(GO:0052773) chitooligosaccharide deacetylase activity(GO:0052790) |
| 0.1 | 105.2 | GO:0003677 | DNA binding(GO:0003677) |
| 0.1 | 0.8 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.0 | 3.5 | GO:0016810 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds(GO:0016810) |
| 0.0 | 0.3 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 1.2 | GO:0008748 | N-ethylmaleimide reductase activity(GO:0008748) reduced coenzyme F420 dehydrogenase activity(GO:0043738) sulfur oxygenase reductase activity(GO:0043826) malolactic enzyme activity(GO:0043883) epoxyqueuosine reductase activity(GO:0052693) |
| 0.0 | 80.7 | GO:0046872 | metal ion binding(GO:0046872) |
| 0.0 | 3.6 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 1.2 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 1.3 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 1.3 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |