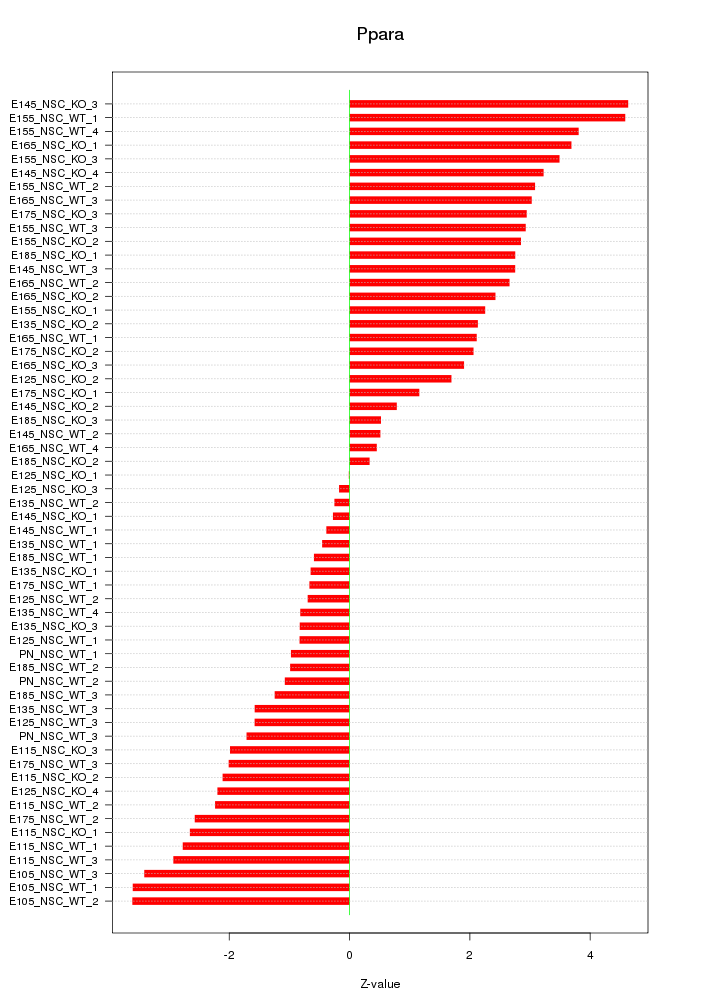
Motif ID: Ppara
Z-value: 2.254
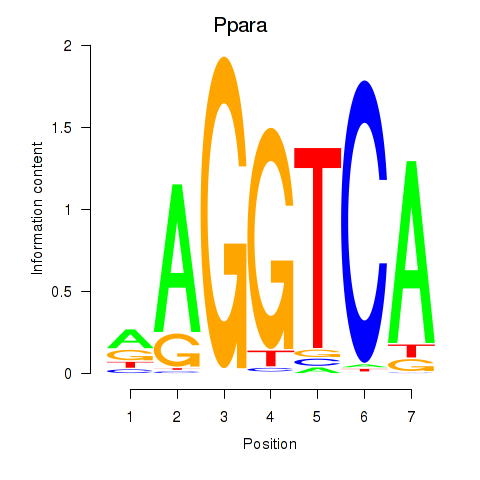
Transcription factors associated with Ppara:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Ppara | ENSMUSG00000022383.7 | Ppara |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 18.9 | 56.8 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 12.5 | 37.5 | GO:0021933 | radial glia guided migration of cerebellar granule cell(GO:0021933) |
| 11.0 | 11.0 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 6.7 | 20.0 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) activation of meiosis involved in egg activation(GO:0060466) |
| 6.5 | 19.5 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 5.1 | 15.4 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 5.1 | 20.3 | GO:1901204 | positive regulation of guanylate cyclase activity(GO:0031284) regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 4.9 | 19.6 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 4.6 | 41.8 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 4.2 | 12.6 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 4.1 | 12.3 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 3.9 | 31.5 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 3.6 | 10.9 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 3.6 | 10.7 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 3.5 | 10.6 | GO:1900736 | regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900736) |
| 3.2 | 19.5 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 3.0 | 24.4 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 3.0 | 8.9 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 2.9 | 8.8 | GO:0010871 | negative regulation of receptor biosynthetic process(GO:0010871) |
| 2.9 | 20.2 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 2.8 | 5.7 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 2.8 | 8.4 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 2.8 | 8.4 | GO:0042560 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 2.7 | 8.2 | GO:0071544 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 2.6 | 10.6 | GO:0055095 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 2.4 | 7.3 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 2.4 | 24.2 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 2.2 | 4.5 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 2.2 | 6.6 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 2.1 | 12.9 | GO:1901524 | regulation of autophagosome maturation(GO:1901096) regulation of macromitophagy(GO:1901524) negative regulation of macromitophagy(GO:1901525) |
| 2.1 | 6.3 | GO:0002447 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 2.0 | 6.1 | GO:0038027 | apolipoprotein A-I-mediated signaling pathway(GO:0038027) |
| 2.0 | 10.1 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 1.9 | 13.6 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 1.8 | 5.5 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleoside transport(GO:0032242) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 1.8 | 7.2 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 1.8 | 7.2 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 1.7 | 15.6 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 1.7 | 8.6 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 1.7 | 8.5 | GO:0030242 | pexophagy(GO:0030242) |
| 1.6 | 13.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 1.6 | 40.4 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 1.6 | 8.0 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 1.6 | 14.4 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 1.6 | 20.8 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 1.6 | 9.4 | GO:0046103 | ADP biosynthetic process(GO:0006172) inosine biosynthetic process(GO:0046103) |
| 1.5 | 6.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 1.5 | 4.5 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 1.5 | 4.5 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 1.5 | 10.4 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 1.4 | 5.7 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 1.4 | 11.2 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 1.4 | 25.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 1.3 | 25.5 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 1.3 | 51.8 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 1.3 | 3.8 | GO:1901228 | regulation of osteoclast proliferation(GO:0090289) negative regulation of bone mineralization involved in bone maturation(GO:1900158) positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 1.2 | 3.7 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 1.2 | 6.0 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 1.2 | 30.1 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 1.2 | 4.7 | GO:1903416 | response to glycoside(GO:1903416) |
| 1.2 | 7.1 | GO:1903056 | regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 1.2 | 4.7 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 1.2 | 4.7 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 1.2 | 5.9 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 1.2 | 37.1 | GO:0061098 | positive regulation of protein tyrosine kinase activity(GO:0061098) |
| 1.1 | 4.5 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 1.1 | 15.8 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 1.1 | 3.4 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 1.1 | 3.3 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) regulation of potassium ion export(GO:1902302) |
| 1.1 | 8.7 | GO:0071732 | cellular response to nitric oxide(GO:0071732) |
| 1.1 | 6.5 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 1.0 | 6.2 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 1.0 | 3.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 1.0 | 7.0 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 1.0 | 2.0 | GO:0042713 | sperm ejaculation(GO:0042713) |
| 1.0 | 5.9 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 1.0 | 3.9 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 1.0 | 6.7 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.9 | 2.8 | GO:1904049 | regulation of spontaneous neurotransmitter secretion(GO:1904048) negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.9 | 2.6 | GO:1900149 | positive regulation of Schwann cell migration(GO:1900149) |
| 0.9 | 9.4 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.8 | 18.2 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.8 | 13.8 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.8 | 18.6 | GO:0030431 | sleep(GO:0030431) |
| 0.8 | 3.9 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.8 | 5.4 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.8 | 28.3 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.8 | 9.9 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.7 | 4.5 | GO:0071472 | cellular response to salt stress(GO:0071472) |
| 0.7 | 3.6 | GO:0070305 | response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) |
| 0.7 | 8.7 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.7 | 6.5 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.7 | 2.1 | GO:0043181 | vacuolar sequestering(GO:0043181) |
| 0.7 | 4.2 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.7 | 4.9 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.7 | 2.8 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.7 | 4.8 | GO:0051036 | regulation of endosome size(GO:0051036) |
| 0.7 | 3.4 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.7 | 2.0 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.7 | 3.3 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.6 | 4.5 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.6 | 1.9 | GO:0032278 | positive regulation of gonadotropin secretion(GO:0032278) |
| 0.6 | 4.5 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.6 | 1.9 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.6 | 12.0 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.6 | 2.5 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.6 | 1.9 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.6 | 8.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.6 | 4.2 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.6 | 1.7 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.6 | 7.5 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.6 | 9.1 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.6 | 7.4 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.6 | 15.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.5 | 11.0 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.5 | 13.5 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.5 | 4.3 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.5 | 4.2 | GO:1902993 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.5 | 3.6 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.5 | 1.5 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.5 | 4.5 | GO:2000058 | regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000058) |
| 0.5 | 1.5 | GO:0071435 | potassium ion export(GO:0071435) |
| 0.5 | 5.4 | GO:1901250 | regulation of lung goblet cell differentiation(GO:1901249) negative regulation of lung goblet cell differentiation(GO:1901250) |
| 0.5 | 14.8 | GO:0051966 | regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.5 | 3.9 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.5 | 7.8 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.5 | 9.3 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.5 | 3.4 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.5 | 7.0 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.5 | 0.9 | GO:0098915 | membrane repolarization during ventricular cardiac muscle cell action potential(GO:0098915) |
| 0.5 | 2.7 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.4 | 6.3 | GO:0043496 | regulation of protein homodimerization activity(GO:0043496) |
| 0.4 | 2.7 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.4 | 15.6 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.4 | 2.2 | GO:0046689 | response to mercury ion(GO:0046689) |
| 0.4 | 3.1 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.4 | 3.9 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.4 | 1.7 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) negative regulation of histone H3-K4 methylation(GO:0051572) |
| 0.4 | 8.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.4 | 8.4 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.4 | 14.2 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.4 | 4.9 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.4 | 10.9 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.4 | 3.2 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) regulation of ribonucleoprotein complex localization(GO:2000197) |
| 0.4 | 1.2 | GO:1903599 | positive regulation of mitophagy(GO:1903599) |
| 0.4 | 7.5 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.4 | 3.1 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.4 | 0.4 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.4 | 3.9 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.4 | 6.6 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.4 | 1.2 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.4 | 0.4 | GO:1903525 | regulation of membrane tubulation(GO:1903525) |
| 0.4 | 4.5 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.4 | 4.0 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.4 | 1.8 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.4 | 0.4 | GO:0002678 | positive regulation of chronic inflammatory response(GO:0002678) |
| 0.4 | 1.1 | GO:0016056 | photoreceptor cell morphogenesis(GO:0008594) rhodopsin mediated signaling pathway(GO:0016056) post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.4 | 1.1 | GO:0021590 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.4 | 5.6 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.4 | 9.5 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.3 | 4.7 | GO:0009072 | aromatic amino acid family metabolic process(GO:0009072) |
| 0.3 | 2.0 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.3 | 7.7 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.3 | 2.0 | GO:0097460 | ferrous iron import into cell(GO:0097460) |
| 0.3 | 6.6 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.3 | 2.3 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.3 | 2.2 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.3 | 3.8 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.3 | 1.3 | GO:0097494 | regulation of vesicle size(GO:0097494) |
| 0.3 | 2.2 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.3 | 5.0 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.3 | 1.2 | GO:0044330 | canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) lactic acid secretion(GO:0046722) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.3 | 1.5 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.3 | 2.1 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) plasma membrane to endosome transport(GO:0048227) regulation of lipoprotein lipase activity(GO:0051004) |
| 0.3 | 1.4 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.3 | 1.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.3 | 5.2 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.3 | 5.7 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.3 | 3.8 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.3 | 1.8 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.3 | 1.0 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.3 | 0.8 | GO:1990000 | negative regulation of protein phosphatase type 2B activity(GO:0032513) amyloid fibril formation(GO:1990000) |
| 0.3 | 1.5 | GO:0019336 | phenol-containing compound catabolic process(GO:0019336) |
| 0.3 | 3.8 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.2 | 1.7 | GO:0060903 | regulation of meiosis I(GO:0060631) positive regulation of meiosis I(GO:0060903) |
| 0.2 | 1.0 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.2 | 0.7 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.2 | 2.6 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.2 | 0.9 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.2 | 2.2 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.2 | 6.5 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.2 | 5.9 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.2 | 1.2 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) amylase secretion(GO:0036394) |
| 0.2 | 2.0 | GO:0051683 | establishment of Golgi localization(GO:0051683) |
| 0.2 | 2.8 | GO:0048520 | positive regulation of behavior(GO:0048520) |
| 0.2 | 1.0 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 0.2 | 2.0 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.2 | 2.8 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.2 | 9.6 | GO:0000045 | autophagosome assembly(GO:0000045) |
| 0.2 | 5.6 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.2 | 1.5 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.2 | 8.3 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.2 | 4.1 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.2 | 23.2 | GO:0030041 | actin filament polymerization(GO:0030041) |
| 0.2 | 2.2 | GO:0006623 | protein targeting to vacuole(GO:0006623) establishment of protein localization to vacuole(GO:0072666) |
| 0.2 | 1.3 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.2 | 2.5 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.2 | 3.6 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.2 | 1.2 | GO:0000022 | mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.2 | 0.7 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.2 | 2.9 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.2 | 1.6 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.2 | 0.6 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.2 | 2.4 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 1.8 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.1 | 2.2 | GO:0001964 | startle response(GO:0001964) |
| 0.1 | 3.0 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.1 | 0.5 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.1 | 6.4 | GO:0045682 | regulation of epidermis development(GO:0045682) |
| 0.1 | 0.3 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.1 | 6.2 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.1 | 1.4 | GO:0097484 | dendrite extension(GO:0097484) |
| 0.1 | 10.6 | GO:0007612 | learning(GO:0007612) |
| 0.1 | 2.6 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.1 | 0.5 | GO:1901798 | positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.1 | 0.4 | GO:0015938 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) |
| 0.1 | 0.5 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.1 | 2.9 | GO:0090004 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 3.6 | GO:0050806 | positive regulation of synaptic transmission(GO:0050806) |
| 0.1 | 1.0 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 2.2 | GO:0001707 | mesoderm formation(GO:0001707) |
| 0.1 | 5.7 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.1 | 7.5 | GO:0010976 | positive regulation of neuron projection development(GO:0010976) |
| 0.1 | 3.4 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.1 | 1.4 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 9.9 | GO:0007286 | spermatid development(GO:0007286) |
| 0.1 | 1.6 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.1 | 0.8 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 1.0 | GO:2000273 | positive regulation of receptor activity(GO:2000273) |
| 0.1 | 0.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 8.2 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 2.2 | GO:0008361 | regulation of cell size(GO:0008361) |
| 0.1 | 3.5 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.1 | 1.1 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 3.5 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) peptidyl-threonine modification(GO:0018210) |
| 0.1 | 1.1 | GO:0071028 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.1 | 5.2 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 0.3 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.1 | 18.4 | GO:0043413 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.1 | 2.7 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 1.0 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.1 | 2.6 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.1 | 0.4 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.1 | 1.8 | GO:0007031 | peroxisome organization(GO:0007031) |
| 0.1 | 1.6 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.1 | 2.0 | GO:0001782 | B cell homeostasis(GO:0001782) |
| 0.1 | 0.5 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 4.0 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 1.4 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.1 | 0.7 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.1 | 1.9 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.1 | 1.7 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.1 | 4.6 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.1 | 1.3 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.1 | 0.5 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.1 | 7.3 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.1 | 1.1 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 0.2 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.1 | 0.3 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.1 | 0.7 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.1 | 1.4 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.1 | 1.2 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.7 | GO:0001659 | temperature homeostasis(GO:0001659) |
| 0.0 | 0.6 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 3.1 | GO:0006073 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 | 1.3 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 1.0 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.4 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 2.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.2 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 1.8 | GO:0016358 | dendrite development(GO:0016358) |
| 0.0 | 0.2 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 1.1 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) |
| 0.0 | 3.9 | GO:0007179 | transforming growth factor beta receptor signaling pathway(GO:0007179) |
| 0.0 | 0.8 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 3.0 | GO:0030534 | adult behavior(GO:0030534) |
| 0.0 | 0.5 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.3 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.6 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 1.3 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.0 | 2.2 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.1 | GO:0002385 | organ or tissue specific immune response(GO:0002251) mucosal immune response(GO:0002385) |
| 0.0 | 0.4 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 2.0 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.5 | GO:0050953 | visual perception(GO:0007601) sensory perception of light stimulus(GO:0050953) |
| 0.0 | 0.7 | GO:0014065 | phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.0 | 2.1 | GO:0010769 | regulation of cell morphogenesis involved in differentiation(GO:0010769) |
| 0.0 | 0.1 | GO:0015858 | nucleoside transport(GO:0015858) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.5 | 56.8 | GO:0097427 | microtubule bundle(GO:0097427) |
| 4.3 | 17.3 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 4.0 | 16.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 3.9 | 38.6 | GO:0044327 | dendritic spine head(GO:0044327) |
| 3.3 | 9.9 | GO:0072534 | perineuronal net(GO:0072534) |
| 3.1 | 15.6 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 2.8 | 8.4 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 2.8 | 8.3 | GO:0032437 | cuticular plate(GO:0032437) |
| 2.5 | 12.7 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 2.5 | 12.6 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 2.1 | 10.6 | GO:0042583 | chromaffin granule(GO:0042583) |
| 2.1 | 37.5 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 1.8 | 20.3 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 1.7 | 10.0 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 1.5 | 7.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 1.4 | 13.0 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 1.3 | 24.9 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 1.3 | 2.6 | GO:0016939 | kinesin II complex(GO:0016939) |
| 1.2 | 3.7 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 1.2 | 24.5 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 1.2 | 9.4 | GO:0001520 | outer dense fiber(GO:0001520) |
| 1.1 | 6.7 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 1.0 | 47.8 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 1.0 | 9.6 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.9 | 2.8 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.9 | 2.7 | GO:1990879 | CST complex(GO:1990879) |
| 0.9 | 1.8 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.9 | 2.6 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.9 | 18.0 | GO:0030673 | axolemma(GO:0030673) |
| 0.8 | 3.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.8 | 12.0 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.7 | 12.5 | GO:0031430 | M band(GO:0031430) |
| 0.7 | 15.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.7 | 21.2 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.7 | 7.4 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.6 | 16.8 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.6 | 5.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.6 | 27.4 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.5 | 6.0 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.5 | 28.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.5 | 4.0 | GO:0042599 | lamellar body(GO:0042599) |
| 0.5 | 1.0 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.5 | 6.4 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.5 | 4.8 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.5 | 6.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.5 | 6.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.4 | 29.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.4 | 3.3 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.4 | 7.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.4 | 9.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.4 | 20.4 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.3 | 4.5 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.3 | 2.9 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.3 | 9.3 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.3 | 13.4 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.3 | 3.6 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.3 | 9.2 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.3 | 5.9 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.3 | 1.7 | GO:0032982 | myosin filament(GO:0032982) |
| 0.3 | 8.9 | GO:0032420 | stereocilium(GO:0032420) |
| 0.3 | 7.1 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.3 | 3.4 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.3 | 7.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.3 | 3.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.3 | 2.0 | GO:0097433 | dense body(GO:0097433) |
| 0.2 | 24.3 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.2 | 30.7 | GO:0005884 | actin filament(GO:0005884) |
| 0.2 | 87.4 | GO:0098794 | postsynapse(GO:0098794) |
| 0.2 | 6.0 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.2 | 2.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.2 | 1.0 | GO:0001652 | granular component(GO:0001652) |
| 0.2 | 24.6 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.2 | 14.8 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.2 | 9.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.2 | 10.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 3.0 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.2 | 6.2 | GO:0005776 | autophagosome(GO:0005776) |
| 0.2 | 6.6 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.2 | 39.7 | GO:0045202 | synapse(GO:0045202) |
| 0.2 | 2.4 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.2 | 2.0 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.2 | 1.1 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.1 | 3.1 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.1 | 21.9 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.1 | 3.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.7 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 3.0 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.1 | 3.0 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.6 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 0.5 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.1 | 3.3 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 1.1 | GO:0019908 | nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.1 | 4.4 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 0.5 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 9.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.9 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 2.5 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 1.7 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 0.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 0.3 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.1 | 0.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 5.2 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 1.8 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 4.9 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.2 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 10.9 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 2.5 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 2.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 4.5 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 2.9 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.3 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 1.9 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.4 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 58.2 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 0.4 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.1 | GO:0042571 | immunoglobulin complex(GO:0019814) immunoglobulin complex, circulating(GO:0042571) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.8 | 20.3 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 6.2 | 18.7 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 5.4 | 32.7 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 4.1 | 12.3 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 3.3 | 10.0 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 3.2 | 19.5 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 3.1 | 24.6 | GO:0103116 | alpha-D-galactofuranose transporter activity(GO:0103116) |
| 3.1 | 9.2 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 3.0 | 14.8 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 2.8 | 8.4 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 2.8 | 8.3 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 2.7 | 8.2 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 2.6 | 15.6 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 2.5 | 7.6 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 2.4 | 9.7 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 2.4 | 7.3 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 2.1 | 12.8 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 2.1 | 12.7 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 2.1 | 10.6 | GO:0055100 | adiponectin binding(GO:0055100) |
| 2.1 | 35.6 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 2.0 | 24.6 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 2.0 | 6.1 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) |
| 2.0 | 6.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 1.9 | 7.8 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 1.9 | 9.6 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 1.8 | 20.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 1.8 | 31.6 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 1.7 | 29.3 | GO:0004890 | GABA-A receptor activity(GO:0004890) GABA receptor activity(GO:0016917) |
| 1.7 | 1.7 | GO:0031755 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 1.5 | 7.5 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 1.4 | 4.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 1.4 | 8.2 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 1.3 | 4.0 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 1.2 | 10.9 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 1.2 | 4.7 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 1.2 | 4.7 | GO:0004954 | icosanoid receptor activity(GO:0004953) prostanoid receptor activity(GO:0004954) prostaglandin receptor activity(GO:0004955) |
| 1.1 | 7.7 | GO:0030274 | LIM domain binding(GO:0030274) |
| 1.1 | 30.9 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 1.1 | 5.4 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 1.1 | 22.7 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 1.1 | 8.4 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 1.0 | 5.2 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 1.0 | 47.9 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 1.0 | 28.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 1.0 | 10.9 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 1.0 | 18.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 1.0 | 1.0 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.9 | 5.7 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.9 | 2.8 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.9 | 4.5 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.9 | 9.4 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.8 | 5.9 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.8 | 4.2 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.8 | 17.7 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.8 | 7.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.8 | 2.5 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.8 | 60.5 | GO:0061733 | peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.8 | 5.6 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.8 | 4.8 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.8 | 3.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.8 | 5.5 | GO:0032795 | G-protein coupled adenosine receptor activity(GO:0001609) heterotrimeric G-protein binding(GO:0032795) |
| 0.7 | 1.5 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.7 | 4.5 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.7 | 3.6 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.7 | 17.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.7 | 5.7 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.7 | 2.1 | GO:0032450 | oligo-1,6-glucosidase activity(GO:0004574) maltose alpha-glucosidase activity(GO:0032450) |
| 0.7 | 4.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.7 | 3.5 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.7 | 4.9 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.7 | 11.1 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.7 | 2.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.7 | 23.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.7 | 2.6 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.6 | 22.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.6 | 15.4 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.6 | 8.0 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.6 | 6.6 | GO:0034943 | acyl-CoA ligase activity(GO:0003996) 3-oxo-2-(2'-pentenyl)cyclopentane-1-octanoic acid CoA ligase activity(GO:0010435) 3-isopropenyl-6-oxoheptanoyl-CoA synthetase activity(GO:0018854) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA synthetase activity(GO:0018855) benzoyl acetate-CoA ligase activity(GO:0018856) 2,4-dichlorobenzoate-CoA ligase activity(GO:0018857) pivalate-CoA ligase activity(GO:0034783) cyclopropanecarboxylate-CoA ligase activity(GO:0034793) adipate-CoA ligase activity(GO:0034796) citronellyl-CoA ligase activity(GO:0034823) mentha-1,3-dione-CoA ligase activity(GO:0034841) thiophene-2-carboxylate-CoA ligase activity(GO:0034842) 2,4,4-trimethylpentanoate-CoA ligase activity(GO:0034865) cis-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034942) trans-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034943) branched-chain acyl-CoA synthetase (ADP-forming) activity(GO:0043759) aryl-CoA synthetase (ADP-forming) activity(GO:0043762) 3-hydroxypropionyl-CoA synthetase activity(GO:0043955) perillic acid:CoA ligase (ADP-forming) activity(GO:0052685) perillic acid:CoA ligase (AMP-forming) activity(GO:0052686) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (ADP-forming) activity(GO:0052687) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (AMP-forming) activity(GO:0052688) pristanate-CoA ligase activity(GO:0070251) malonyl-CoA synthetase activity(GO:0090409) |
| 0.6 | 5.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.6 | 5.7 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.6 | 8.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.5 | 24.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.5 | 1.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.5 | 3.8 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.5 | 15.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.5 | 5.9 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.5 | 7.3 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.5 | 1.4 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.5 | 10.1 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.5 | 1.4 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.5 | 17.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.5 | 8.8 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.5 | 1.8 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.4 | 3.6 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.4 | 6.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.4 | 15.9 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.4 | 1.3 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.4 | 3.8 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.4 | 2.9 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.4 | 2.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.4 | 7.7 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.4 | 4.8 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.4 | 4.4 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.4 | 1.2 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.4 | 3.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.4 | 4.7 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.4 | 1.2 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.4 | 12.6 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.4 | 1.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.4 | 6.7 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.4 | 4.3 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.3 | 3.8 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.3 | 2.0 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.3 | 9.8 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.3 | 2.0 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.3 | 6.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.3 | 1.9 | GO:0019841 | retinol binding(GO:0019841) |
| 0.3 | 6.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.3 | 1.3 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.3 | 9.1 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.3 | 1.9 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.3 | 2.2 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.3 | 2.7 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.3 | 7.1 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.3 | 8.7 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.3 | 2.0 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.3 | 1.1 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.3 | 1.9 | GO:0016004 | phospholipase activator activity(GO:0016004) lipase activator activity(GO:0060229) |
| 0.3 | 1.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.3 | 3.3 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.2 | 1.7 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.2 | 3.7 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.2 | 1.0 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) |
| 0.2 | 2.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.2 | 3.3 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.2 | 3.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.2 | 15.3 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.2 | 1.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.2 | 2.7 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.2 | 4.2 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.2 | 6.4 | GO:0034930 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) heparan sulfate 2-O-sulfotransferase activity(GO:0004394) HNK-1 sulfotransferase activity(GO:0016232) heparan sulfate 6-O-sulfotransferase activity(GO:0017095) trans-9R,10R-dihydrodiolphenanthrene sulfotransferase activity(GO:0018721) 1-phenanthrol sulfotransferase activity(GO:0018722) 3-phenanthrol sulfotransferase activity(GO:0018723) 4-phenanthrol sulfotransferase activity(GO:0018724) trans-3,4-dihydrodiolphenanthrene sulfotransferase activity(GO:0018725) 9-phenanthrol sulfotransferase activity(GO:0018726) 2-phenanthrol sulfotransferase activity(GO:0018727) phenanthrol sulfotransferase activity(GO:0019111) 1-hydroxypyrene sulfotransferase activity(GO:0034930) proteoglycan sulfotransferase activity(GO:0050698) cholesterol sulfotransferase activity(GO:0051922) hydroxyjasmonate sulfotransferase activity(GO:0080131) |
| 0.2 | 4.4 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.2 | 1.2 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.2 | 2.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 7.4 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.2 | 0.7 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.2 | 5.1 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.2 | 2.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.2 | 5.7 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.2 | 5.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.2 | 0.6 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.1 | 8.3 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 8.5 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 1.0 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 22.1 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 4.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.5 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.1 | 0.7 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 0.8 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.1 | 6.7 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 2.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 2.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 1.4 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 1.6 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 4.7 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 8.0 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.1 | 1.0 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 1.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.1 | 1.9 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 0.6 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 0.9 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 0.3 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 8.4 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 5.8 | GO:0019209 | kinase activator activity(GO:0019209) |
| 0.1 | 0.4 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.1 | 1.0 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 2.7 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 6.5 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 2.6 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 3.6 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 1.7 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.1 | 1.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.8 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 0.7 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 15.1 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.1 | 5.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.3 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 6.3 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.1 | 0.2 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.1 | 0.4 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 2.5 | GO:0008748 | N-ethylmaleimide reductase activity(GO:0008748) reduced coenzyme F420 dehydrogenase activity(GO:0043738) sulfur oxygenase reductase activity(GO:0043826) malolactic enzyme activity(GO:0043883) epoxyqueuosine reductase activity(GO:0052693) |
| 0.1 | 3.3 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 5.4 | GO:0043851 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) rRNA (uridine-2'-O-)-methyltransferase activity(GO:0008650) rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) selenocysteine methyltransferase activity(GO:0016205) rRNA (adenine) methyltransferase activity(GO:0016433) rRNA (cytosine) methyltransferase activity(GO:0016434) rRNA (guanine) methyltransferase activity(GO:0016435) 1-phenanthrol methyltransferase activity(GO:0018707) protein-arginine N5-methyltransferase activity(GO:0019702) dimethylarsinite methyltransferase activity(GO:0034541) 4,5-dihydroxybenzo(a)pyrene methyltransferase activity(GO:0034807) 1-hydroxypyrene methyltransferase activity(GO:0034931) 1-hydroxy-6-methoxypyrene methyltransferase activity(GO:0034933) demethylmenaquinone methyltransferase activity(GO:0043770) cobalt-precorrin-6B C5-methyltransferase activity(GO:0043776) cobalt-precorrin-7 C15-methyltransferase activity(GO:0043777) cobalt-precorrin-5B C1-methyltransferase activity(GO:0043780) cobalt-precorrin-3 C17-methyltransferase activity(GO:0043782) dimethylamine methyltransferase activity(GO:0043791) hydroxyneurosporene-O-methyltransferase activity(GO:0043803) tRNA (adenine-57, 58-N(1)-) methyltransferase activity(GO:0043827) methylamine-specific methylcobalamin:coenzyme M methyltransferase activity(GO:0043833) trimethylamine methyltransferase activity(GO:0043834) methanol-specific methylcobalamin:coenzyme M methyltransferase activity(GO:0043851) monomethylamine methyltransferase activity(GO:0043852) P-methyltransferase activity(GO:0051994) Se-methyltransferase activity(GO:0051995) 2-phytyl-1,4-naphthoquinone methyltransferase activity(GO:0052624) tRNA (uracil-2'-O-)-methyltransferase activity(GO:0052665) tRNA (cytosine-2'-O-)-methyltransferase activity(GO:0052666) phosphomethylethanolamine N-methyltransferase activity(GO:0052667) tRNA (cytosine-3-)-methyltransferase activity(GO:0052735) rRNA (cytosine-2'-O-)-methyltransferase activity(GO:0070677) rRNA (cytosine-N4-)-methyltransferase activity(GO:0071424) trihydroxyferuloyl spermidine O-methyltransferase activity(GO:0080012) |
| 0.1 | 2.0 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 1.1 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 1.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 2.8 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.1 | 1.0 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 1.8 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 0.5 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.1 | 0.2 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.1 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 2.0 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.4 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.7 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 2.8 | GO:0000976 | transcription regulatory region sequence-specific DNA binding(GO:0000976) |
| 0.0 | 2.1 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 3.9 | GO:0016879 | ligase activity, forming carbon-nitrogen bonds(GO:0016879) |
| 0.0 | 0.4 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.0 | 3.2 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.3 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.0 | 0.6 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 10.3 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 1.9 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.0 | 0.3 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.4 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 1.3 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.2 | GO:0004889 | acetylcholine-activated cation-selective channel activity(GO:0004889) |
| 0.0 | 2.4 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 1.7 | GO:0008514 | organic anion transmembrane transporter activity(GO:0008514) |
| 0.0 | 0.3 | GO:0050253 | prenylcysteine methylesterase activity(GO:0010296) 1-oxa-2-oxocycloheptane lactonase activity(GO:0018731) sulfolactone hydrolase activity(GO:0018732) butyrolactone hydrolase activity(GO:0018734) endosulfan lactone lactonase activity(GO:0034892) L-ascorbate 6-phosphate lactonase activity(GO:0035460) Ser-tRNA(Thr) hydrolase activity(GO:0043905) Ala-tRNA(Pro) hydrolase activity(GO:0043906) Cys-tRNA(Pro) hydrolase activity(GO:0043907) Ser(Gly)-tRNA(Ala) hydrolase activity(GO:0043908) all-trans-retinyl-palmitate hydrolase, all-trans-retinol forming activity(GO:0047376) retinyl-palmitate esterase activity(GO:0050253) mannosyl-oligosaccharide 1,6-alpha-mannosidase activity(GO:0052767) mannosyl-oligosaccharide 1,3-alpha-mannosidase activity(GO:0052768) methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.0 | 0.7 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 1.3 | GO:0017137 | Rab GTPase binding(GO:0017137) |