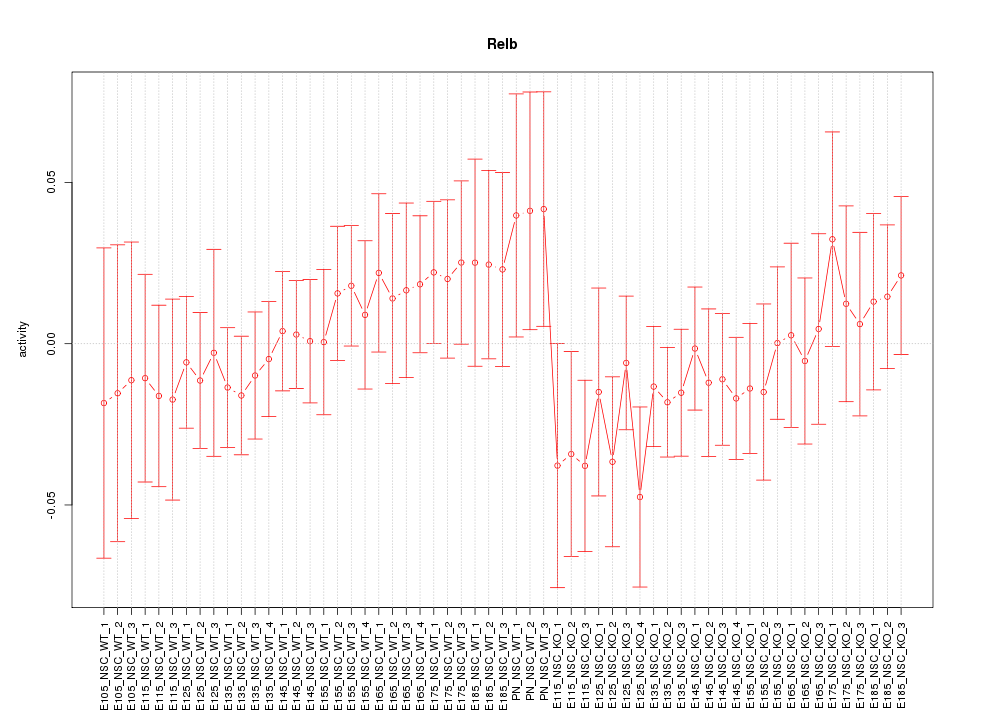
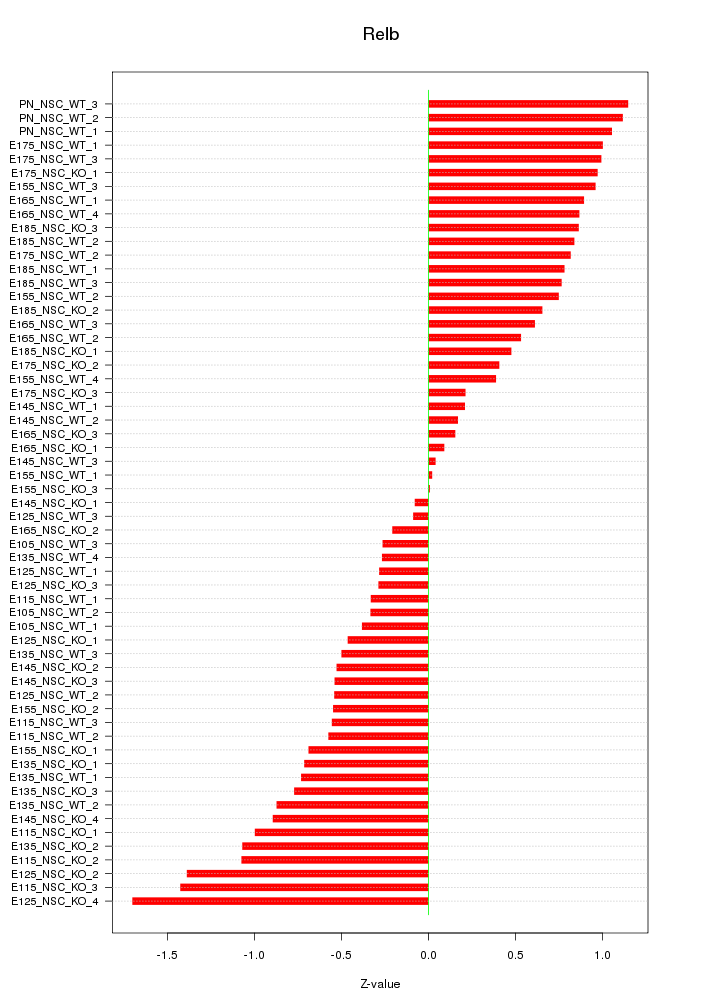
Motif ID: Relb
Z-value: 0.732
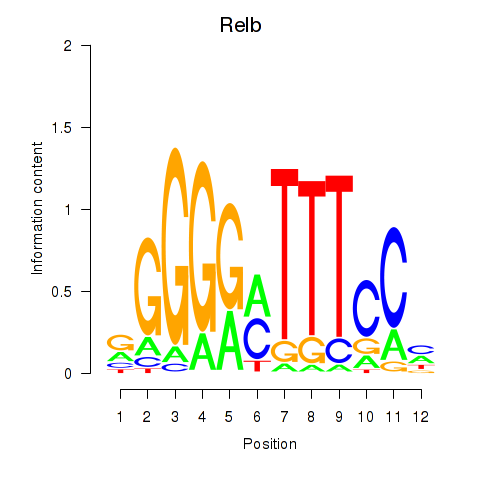
Transcription factors associated with Relb:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Relb | ENSMUSG00000002983.10 | Relb |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Relb | mm10_v2_chr7_-_19629355_19629451 | 0.66 | 1.6e-08 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 8.6 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 1.3 | 9.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.9 | 2.6 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.8 | 5.6 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.8 | 9.2 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.7 | 5.5 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.4 | 1.8 | GO:0009597 | detection of virus(GO:0009597) |
| 0.4 | 2.3 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.4 | 11.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.3 | 1.4 | GO:0061428 | positive regulation of oocyte development(GO:0060282) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.3 | 1.1 | GO:0036015 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.2 | 1.4 | GO:0071499 | response to laminar fluid shear stress(GO:0034616) cellular response to laminar fluid shear stress(GO:0071499) |
| 0.2 | 2.2 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.2 | 4.7 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.2 | 1.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.2 | 1.8 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.1 | 0.9 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.1 | 4.7 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.1 | 0.8 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.1 | 0.5 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 2.6 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 0.5 | GO:0030836 | positive regulation of actin filament depolymerization(GO:0030836) |
| 0.0 | 0.6 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.3 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.1 | GO:0051712 | positive regulation of killing of cells of other organism(GO:0051712) |
| 0.0 | 4.5 | GO:0042060 | wound healing(GO:0042060) |
| 0.0 | 0.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 9.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.9 | 2.6 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.4 | 5.6 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.3 | 2.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.2 | 1.4 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.2 | 2.6 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 0.6 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 0.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 4.7 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 4.9 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 8.6 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.8 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 1.8 | GO:0005923 | bicellular tight junction(GO:0005923) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.6 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 1.1 | 4.5 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 1.1 | 8.6 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.3 | 1.4 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) tubulin deacetylase activity(GO:0042903) |
| 0.3 | 1.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.2 | 9.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 1.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.1 | 9.2 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.1 | 4.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.5 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 2.6 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 1.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 4.7 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 3.8 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 0.3 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 2.1 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 2.7 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 1.8 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 1.8 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |