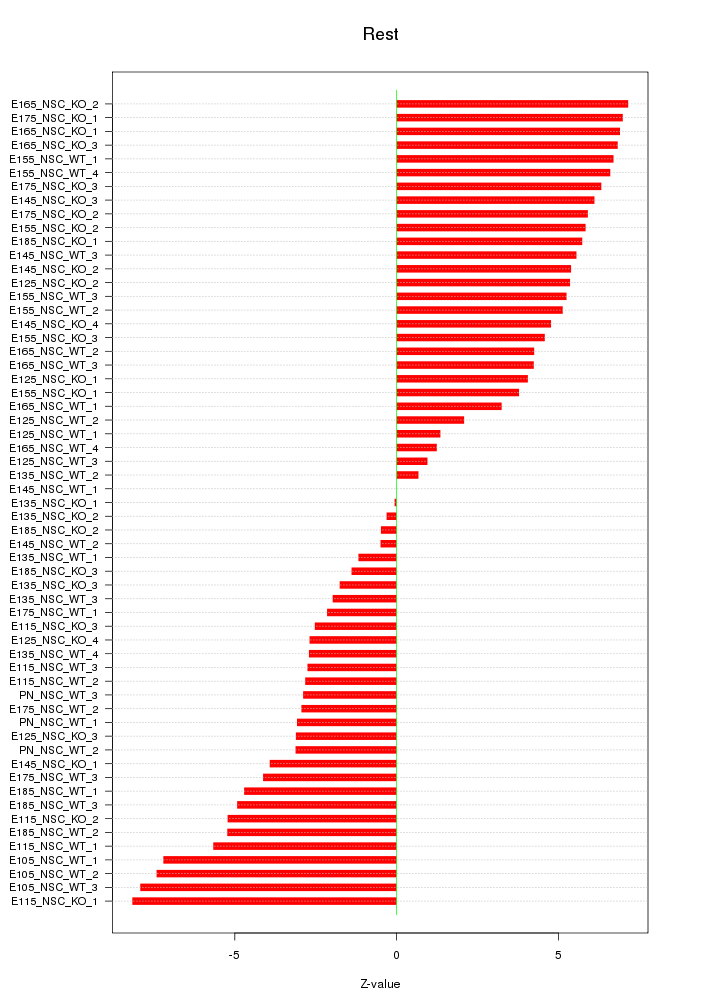
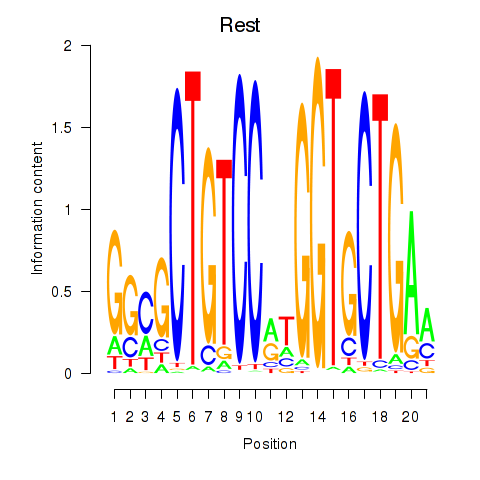
Motif ID: Rest
Z-value: 4.575
Transcription factors associated with Rest:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Rest | ENSMUSG00000029249.9 | Rest |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rest | mm10_v2_chr5_+_77266196_77266232 | -0.70 | 6.7e-10 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 33.5 | 133.8 | GO:0021586 | pons maturation(GO:0021586) |
| 18.2 | 164.0 | GO:0060373 | regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 16.4 | 49.3 | GO:0030827 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 15.9 | 47.7 | GO:1900736 | regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900736) |
| 15.5 | 46.5 | GO:0060084 | conditioned taste aversion(GO:0001661) regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) circadian sleep/wake cycle, REM sleep(GO:0042747) synaptic transmission involved in micturition(GO:0060084) |
| 15.1 | 75.4 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 13.9 | 41.6 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 13.3 | 133.4 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 11.5 | 34.5 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) regulation of opioid receptor signaling pathway(GO:2000474) |
| 11.2 | 101.0 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 7.4 | 37.2 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 7.4 | 44.2 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 6.5 | 39.2 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 5.4 | 32.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 5.0 | 19.9 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 4.8 | 52.7 | GO:0044331 | cell-cell adhesion mediated by cadherin(GO:0044331) |
| 4.6 | 92.6 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 3.9 | 19.6 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 3.5 | 35.3 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 2.9 | 61.8 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 2.9 | 55.9 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 2.7 | 66.4 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 2.6 | 31.4 | GO:0043084 | penile erection(GO:0043084) |
| 2.5 | 37.2 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 2.4 | 26.4 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 2.4 | 56.5 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 2.1 | 8.6 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 2.0 | 10.2 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 1.9 | 26.8 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 1.9 | 83.5 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 1.8 | 5.4 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 1.8 | 24.9 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 1.4 | 28.8 | GO:0001504 | neurotransmitter uptake(GO:0001504) |
| 1.4 | 44.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 1.2 | 25.5 | GO:0071625 | vocalization behavior(GO:0071625) |
| 1.1 | 14.0 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.9 | 6.4 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.9 | 6.2 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.7 | 47.5 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.7 | 32.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.5 | 1.5 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.5 | 3.0 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.5 | 6.4 | GO:0007614 | short-term memory(GO:0007614) |
| 0.5 | 32.5 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.4 | 9.5 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.4 | 37.3 | GO:0099643 | neurotransmitter secretion(GO:0007269) presynaptic process involved in chemical synaptic transmission(GO:0099531) signal release from synapse(GO:0099643) |
| 0.4 | 4.7 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.3 | 34.1 | GO:0014065 | phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.2 | 53.1 | GO:0060828 | regulation of canonical Wnt signaling pathway(GO:0060828) |
| 0.2 | 11.6 | GO:0051341 | regulation of oxidoreductase activity(GO:0051341) |
| 0.2 | 140.2 | GO:0030334 | regulation of cell migration(GO:0030334) |
| 0.2 | 23.6 | GO:0007286 | spermatid development(GO:0007286) |
| 0.2 | 7.3 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.2 | 13.3 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.1 | 16.2 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
| 0.1 | 7.5 | GO:0006906 | vesicle fusion(GO:0006906) |
| 0.0 | 6.3 | GO:0051480 | regulation of cytosolic calcium ion concentration(GO:0051480) |
| 0.0 | 22.4 | GO:0009607 | response to biotic stimulus(GO:0009607) |
| 0.0 | 1.1 | GO:0015991 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 2.4 | GO:0006639 | neutral lipid metabolic process(GO:0006638) acylglycerol metabolic process(GO:0006639) |
| 0.0 | 12.6 | GO:0031175 | neuron projection development(GO:0031175) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 22.5 | 67.5 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 16.6 | 66.4 | GO:0044307 | dendritic branch(GO:0044307) |
| 10.3 | 164.0 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 9.5 | 47.7 | GO:0042583 | chromaffin granule(GO:0042583) |
| 9.4 | 75.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 8.4 | 92.6 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 5.9 | 88.7 | GO:0031045 | dense core granule(GO:0031045) |
| 5.4 | 32.4 | GO:0008091 | spectrin(GO:0008091) |
| 5.1 | 25.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 4.9 | 49.3 | GO:0044327 | dendritic spine head(GO:0044327) |
| 4.9 | 19.6 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 4.6 | 209.8 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 4.2 | 46.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 3.9 | 11.8 | GO:0002141 | stereocilia ankle link(GO:0002141) |
| 2.6 | 136.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 2.3 | 66.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 2.2 | 85.4 | GO:0097440 | apical dendrite(GO:0097440) |
| 2.2 | 69.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 1.4 | 56.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.7 | 31.4 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.6 | 37.2 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.4 | 16.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.3 | 49.0 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.3 | 169.7 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.3 | 46.4 | GO:0030141 | secretory granule(GO:0030141) |
| 0.2 | 9.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.2 | 12.3 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.2 | 13.3 | GO:0044297 | cell body(GO:0044297) |
| 0.1 | 47.5 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.1 | 179.4 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.1 | 19.9 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 1.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 142.7 | GO:0005886 | plasma membrane(GO:0005886) |
| 0.1 | 148.0 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.1 | 6.3 | GO:0045202 | synapse(GO:0045202) |
| 0.1 | 3.7 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 83.8 | GO:0005634 | nucleus(GO:0005634) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 26.7 | 133.4 | GO:0033691 | sialic acid binding(GO:0033691) |
| 23.4 | 164.0 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 9.3 | 55.9 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 8.9 | 26.8 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
| 6.0 | 66.4 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 4.5 | 44.7 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 4.2 | 46.5 | GO:0004889 | acetylcholine-activated cation-selective channel activity(GO:0004889) |
| 3.1 | 37.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 3.0 | 171.0 | GO:0005179 | hormone activity(GO:0005179) |
| 2.9 | 85.4 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 2.9 | 67.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 2.5 | 54.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 1.8 | 66.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 1.8 | 19.9 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 1.8 | 47.5 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 1.5 | 28.8 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 1.4 | 56.5 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 1.2 | 8.6 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 1.2 | 61.4 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 1.2 | 75.4 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 1.1 | 52.7 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.9 | 5.4 | GO:0015464 | acetylcholine receptor activity(GO:0015464) |
| 0.9 | 26.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.9 | 9.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.7 | 32.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.6 | 16.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.5 | 23.6 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.5 | 56.0 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.5 | 37.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.4 | 10.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.4 | 47.5 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.4 | 7.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.3 | 14.0 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
| 0.2 | 19.6 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.2 | 89.7 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.2 | 3.0 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.2 | 13.3 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.2 | 11.6 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.1 | 32.3 | GO:0003729 | mRNA binding(GO:0003729) |
| 0.1 | 36.0 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.1 | 2.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 9.8 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 468.0 | GO:0003674 | molecular_function(GO:0003674) |