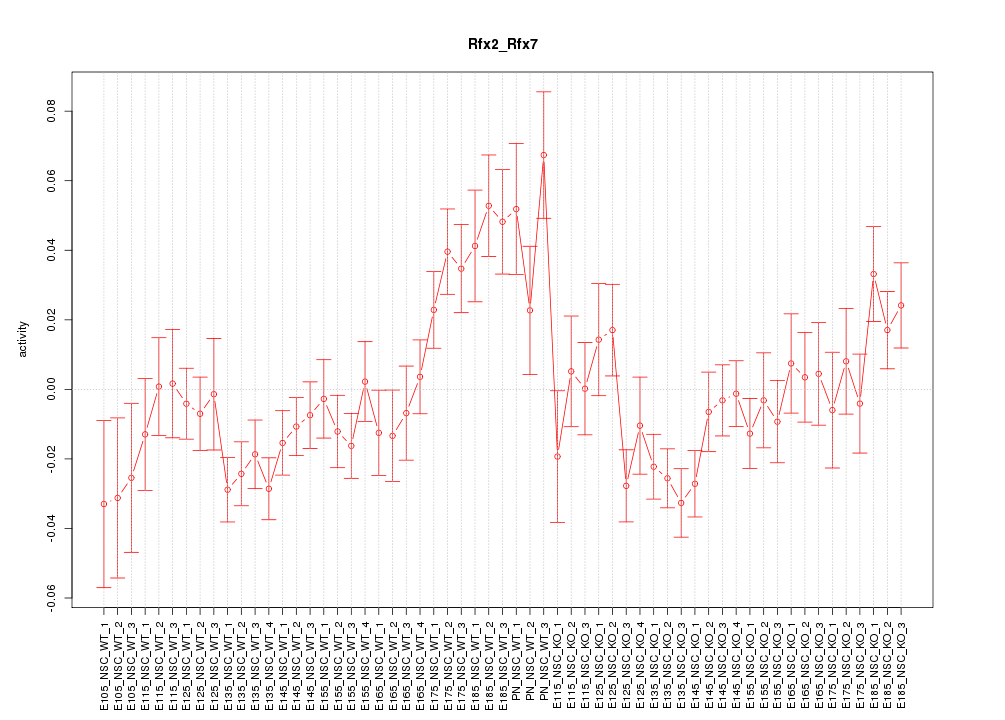
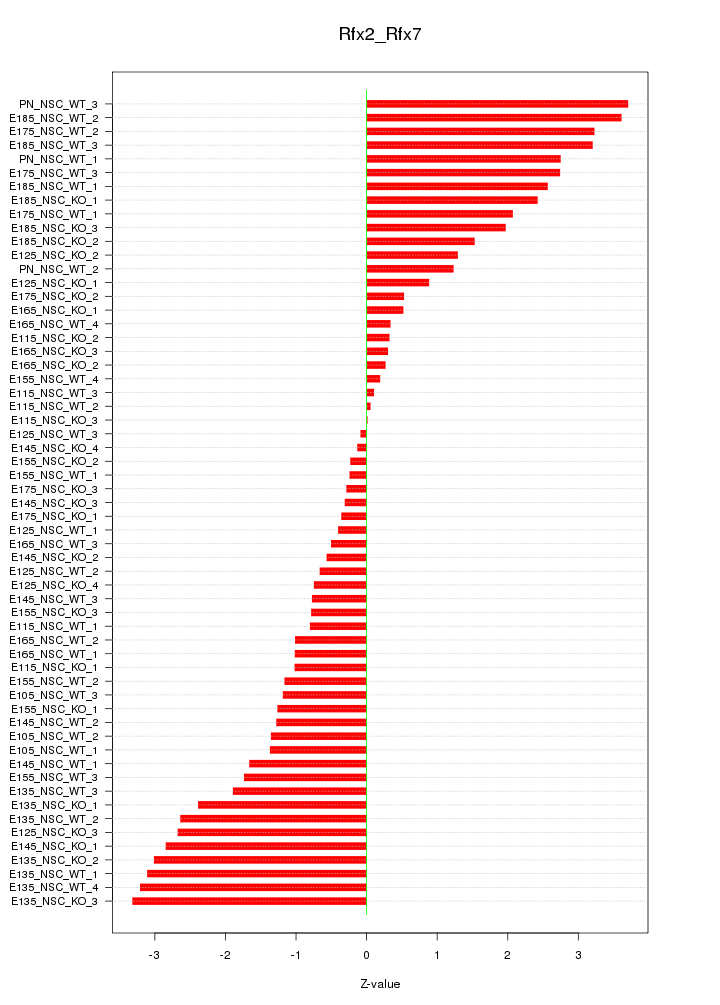
Motif ID: Rfx2_Rfx7
Z-value: 1.770
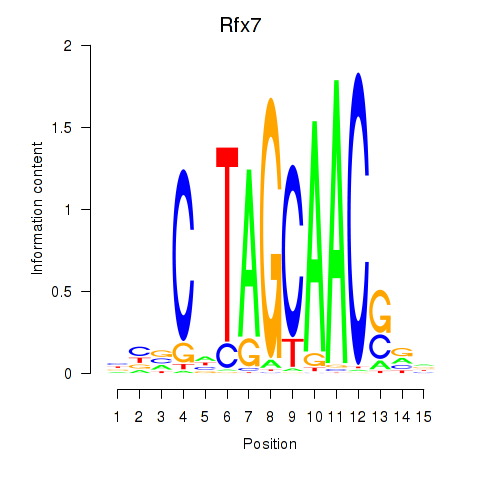
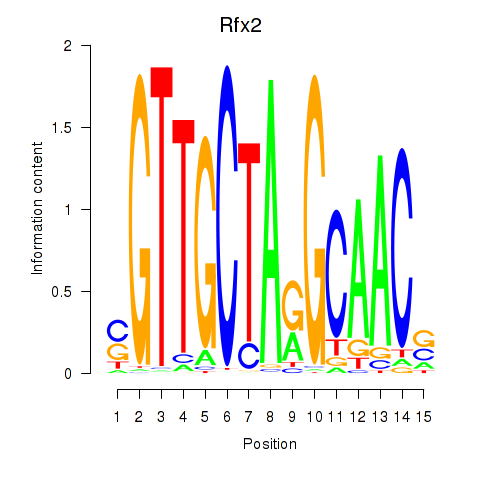
Transcription factors associated with Rfx2_Rfx7:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Rfx2 | ENSMUSG00000024206.8 | Rfx2 |
| Rfx7 | ENSMUSG00000037674.9 | Rfx7 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rfx7 | mm10_v2_chr9_+_72532609_72532828 | 0.28 | 2.9e-02 | Click! |
| Rfx2 | mm10_v2_chr17_-_56830916_56831008 | 0.19 | 1.6e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.4 | 31.9 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 3.2 | 9.7 | GO:2000314 | negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 1.8 | 7.4 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 1.6 | 14.8 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 1.5 | 7.6 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 1.2 | 3.5 | GO:0021893 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 1.0 | 4.2 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.9 | 3.5 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.9 | 3.5 | GO:1902512 | positive regulation of apoptotic DNA fragmentation(GO:1902512) positive regulation of DNA catabolic process(GO:1903626) |
| 0.8 | 6.5 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.8 | 6.4 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.7 | 6.0 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.7 | 4.5 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.7 | 6.5 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.6 | 2.6 | GO:0018307 | enzyme active site formation(GO:0018307) |
| 0.6 | 1.1 | GO:2000282 | regulation of cellular amino acid biosynthetic process(GO:2000282) |
| 0.6 | 5.1 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.6 | 1.7 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.5 | 13.0 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.5 | 3.8 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.5 | 2.2 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.5 | 2.7 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.5 | 1.8 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.5 | 0.9 | GO:1903441 | protein localization to ciliary membrane(GO:1903441) |
| 0.4 | 2.2 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.4 | 2.2 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.4 | 7.4 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.4 | 1.2 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.4 | 4.0 | GO:0046036 | GTP biosynthetic process(GO:0006183) CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.4 | 1.5 | GO:0097393 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.4 | 2.2 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.4 | 3.6 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.4 | 3.2 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.3 | 2.0 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.3 | 1.3 | GO:0061428 | positive regulation of oocyte development(GO:0060282) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.3 | 2.6 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.3 | 4.0 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.3 | 2.0 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.3 | 1.7 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.3 | 2.5 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.3 | 1.6 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.3 | 1.6 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.3 | 0.8 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.2 | 3.4 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.2 | 1.2 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.2 | 1.6 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.2 | 1.9 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.2 | 1.1 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.2 | 0.7 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.2 | 3.4 | GO:0032620 | interleukin-17 production(GO:0032620) |
| 0.2 | 0.2 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.2 | 0.8 | GO:1905049 | negative regulation of metalloendopeptidase activity(GO:1904684) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.2 | 1.1 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.2 | 0.6 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.2 | 4.7 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.2 | 2.7 | GO:0003341 | cilium movement(GO:0003341) |
| 0.2 | 3.0 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 0.2 | 0.5 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.2 | 2.0 | GO:0036376 | sodium ion export from cell(GO:0036376) |
| 0.2 | 0.5 | GO:0060854 | patterning of lymph vessels(GO:0060854) |
| 0.2 | 0.9 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.2 | 2.9 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.2 | 5.6 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.2 | 18.1 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.2 | 2.4 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.2 | 0.6 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.2 | 4.2 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.2 | 1.5 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.2 | 0.6 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 | 0.3 | GO:0090148 | membrane fission(GO:0090148) |
| 0.1 | 16.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.1 | 2.3 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.1 | 1.1 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.1 | 0.8 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 1.7 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 0.5 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.1 | 0.5 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 4.2 | GO:1901381 | positive regulation of potassium ion transmembrane transport(GO:1901381) |
| 0.1 | 0.6 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.1 | 0.6 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 6.9 | GO:0034620 | cellular response to unfolded protein(GO:0034620) |
| 0.1 | 5.6 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.1 | 0.4 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.1 | 0.5 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.5 | GO:2001185 | regulation of CD8-positive, alpha-beta T cell activation(GO:2001185) |
| 0.1 | 0.4 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.1 | 0.3 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.1 | 0.3 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.1 | 1.9 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.1 | 0.4 | GO:0043383 | negative T cell selection(GO:0043383) negative thymic T cell selection(GO:0045060) |
| 0.1 | 0.5 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.1 | 0.8 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.1 | 0.9 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 0.7 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 1.2 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.1 | 0.3 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.1 | 0.1 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.1 | 3.7 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.1 | 11.4 | GO:0051321 | meiotic cell cycle(GO:0051321) |
| 0.1 | 0.3 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 0.7 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.1 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.3 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.1 | 0.2 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.1 | 0.2 | GO:1902224 | cellular ketone body metabolic process(GO:0046950) ketone body metabolic process(GO:1902224) |
| 0.1 | 0.8 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.1 | 1.2 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.5 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 1.1 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.0 | 2.2 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.4 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 7.7 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.5 | GO:0046519 | sphingoid metabolic process(GO:0046519) |
| 0.0 | 1.0 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 1.5 | GO:0060351 | cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.0 | 0.3 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 1.3 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.3 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 1.8 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 2.4 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.0 | 1.2 | GO:1902850 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.0 | 3.6 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 0.3 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 | 1.0 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.2 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.0 | 0.2 | GO:0043084 | penile erection(GO:0043084) |
| 0.0 | 1.6 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.5 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 0.7 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.6 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.8 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.4 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 2.1 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.0 | 2.5 | GO:0098780 | mitophagy in response to mitochondrial depolarization(GO:0098779) response to mitochondrial depolarisation(GO:0098780) |
| 0.0 | 0.7 | GO:0048524 | positive regulation of viral process(GO:0048524) |
| 0.0 | 0.2 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.5 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.1 | GO:0006298 | mismatch repair(GO:0006298) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.5 | GO:0036156 | inner dynein arm(GO:0036156) |
| 2.1 | 31.9 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 1.5 | 6.0 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 1.4 | 27.4 | GO:0097228 | sperm principal piece(GO:0097228) |
| 1.3 | 7.7 | GO:0036157 | axonemal dynein complex(GO:0005858) outer dynein arm(GO:0036157) |
| 0.9 | 3.5 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.7 | 9.2 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.6 | 3.5 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.6 | 1.7 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.5 | 4.4 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.5 | 1.9 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.4 | 4.0 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.4 | 1.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.4 | 1.5 | GO:1990421 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 0.3 | 7.6 | GO:0005605 | basal lamina(GO:0005605) |
| 0.3 | 8.8 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.3 | 3.4 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.2 | 3.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 3.6 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.2 | 1.2 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.2 | 4.2 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.2 | 16.4 | GO:0031514 | motile cilium(GO:0031514) |
| 0.2 | 0.5 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.2 | 19.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 2.9 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 0.4 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 2.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 1.6 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 1.3 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 2.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 2.5 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 3.0 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 1.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 3.1 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 2.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 1.5 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 1.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.5 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 2.2 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 0.8 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 0.6 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 0.8 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 3.4 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.1 | 5.2 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.7 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 1.0 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.5 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.0 | 0.5 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.2 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 1.3 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 2.1 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 1.9 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 2.2 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 2.4 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 4.9 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.6 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.0 | 0.5 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.2 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 1.5 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.6 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.7 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.4 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 1.8 | GO:0005773 | vacuole(GO:0005773) |
| 0.0 | 0.1 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 4.5 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 0.6 | GO:0005811 | lipid particle(GO:0005811) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 31.9 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) calcium-independent phospholipase A2 activity(GO:0047499) |
| 1.8 | 5.4 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 1.2 | 6.0 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.9 | 7.6 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.9 | 18.0 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.7 | 2.9 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.7 | 2.1 | GO:0043912 | D-lysine oxidase activity(GO:0043912) |
| 0.7 | 3.5 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.6 | 1.8 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.6 | 4.7 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.5 | 2.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.5 | 2.6 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.5 | 2.9 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.4 | 2.2 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.4 | 1.6 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.4 | 1.9 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.4 | 1.1 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.3 | 1.3 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) tubulin deacetylase activity(GO:0042903) |
| 0.3 | 1.6 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.3 | 0.9 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.3 | 2.0 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.3 | 3.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.2 | 1.5 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.2 | 3.4 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.2 | 2.2 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.2 | 4.0 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.2 | 0.7 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.2 | 1.5 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.2 | 1.0 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.2 | 1.8 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 8.7 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.2 | 2.0 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.2 | 1.7 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.2 | 3.4 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.2 | 10.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.2 | 10.7 | GO:0003774 | motor activity(GO:0003774) |
| 0.2 | 2.3 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.2 | 2.4 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.2 | 0.5 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.2 | 1.9 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.2 | 0.6 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 2.5 | GO:0050811 | GABA-A receptor activity(GO:0004890) GABA receptor binding(GO:0050811) |
| 0.1 | 3.4 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 19.2 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 0.5 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.1 | 2.5 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.1 | 0.7 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 0.3 | GO:0005119 | smoothened binding(GO:0005119) hedgehog family protein binding(GO:0097108) |
| 0.1 | 1.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 1.7 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 1.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.3 | GO:0019002 | GMP binding(GO:0019002) |
| 0.1 | 1.9 | GO:0035380 | C-3 sterol dehydrogenase (C-4 sterol decarboxylase) activity(GO:0000252) mevaldate reductase activity(GO:0004495) gluconate dehydrogenase activity(GO:0008875) epoxide dehydrogenase activity(GO:0018451) 5-exo-hydroxycamphor dehydrogenase activity(GO:0018452) 2-hydroxytetrahydrofuran dehydrogenase activity(GO:0018453) acetoin dehydrogenase activity(GO:0019152) phenylcoumaran benzylic ether reductase activity(GO:0032442) D-xylose:NADP reductase activity(GO:0032866) L-arabinose:NADP reductase activity(GO:0032867) D-arabinitol dehydrogenase, D-ribulose forming (NADP+) activity(GO:0033709) (R)-(-)-1,2,3,4-tetrahydronaphthol dehydrogenase activity(GO:0034831) 3-hydroxymenthone dehydrogenase activity(GO:0034840) very long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0035380) dihydrotestosterone 17-beta-dehydrogenase activity(GO:0035410) (R)-2-hydroxyisocaproate dehydrogenase activity(GO:0043713) L-arabinose 1-dehydrogenase (NADP+) activity(GO:0044103) L-xylulose reductase (NAD+) activity(GO:0044105) 3-ketoglucose-reductase activity(GO:0048258) D-arabinitol dehydrogenase, D-xylulose forming (NADP+) activity(GO:0052677) |
| 0.1 | 0.7 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.7 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 1.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 4.7 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 1.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.9 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 1.0 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 3.5 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 0.7 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.4 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.1 | 0.2 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.1 | 1.9 | GO:0034945 | dihydrolipoamide branched chain acyltransferase activity(GO:0004147) palmitoleoyl [acyl-carrier-protein]-dependent acyltransferase activity(GO:0008951) serine O-acyltransferase activity(GO:0016412) O-succinyltransferase activity(GO:0016750) sinapoyltransferase activity(GO:0016752) O-sinapoyltransferase activity(GO:0016753) peptidyl-lysine N6-myristoyltransferase activity(GO:0018030) peptidyl-lysine N6-palmitoyltransferase activity(GO:0018031) benzoyl acetate-CoA thiolase activity(GO:0018711) 3-hydroxybutyryl-CoA thiolase activity(GO:0018712) 3-ketopimelyl-CoA thiolase activity(GO:0018713) N-palmitoyltransferase activity(GO:0019105) acyl-CoA N-acyltransferase activity(GO:0019186) protein-cysteine S-myristoyltransferase activity(GO:0019705) glucosaminyl-phosphotidylinositol O-acyltransferase activity(GO:0032216) ergosterol O-acyltransferase activity(GO:0034737) lanosterol O-acyltransferase activity(GO:0034738) naphthyl-2-oxomethyl-succinyl-CoA succinyl transferase activity(GO:0034848) 2,4,4-trimethyl-3-oxopentanoyl-CoA 2-C-propanoyl transferase activity(GO:0034851) 2-methylhexanoyl-CoA C-acetyltransferase activity(GO:0034915) butyryl-CoA 2-C-propionyltransferase activity(GO:0034919) 2,6-dimethyl-5-methylene-3-oxo-heptanoyl-CoA C-acetyltransferase activity(GO:0034945) L-2-aminoadipate N-acetyltransferase activity(GO:0043741) keto acid formate lyase activity(GO:0043806) azetidine-2-carboxylic acid acetyltransferase activity(GO:0046941) peptidyl-lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0052858) acetyl-CoA:L-lysine N6-acetyltransferase(GO:0090595) |
| 0.1 | 2.0 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.1 | 2.1 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.7 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 1.1 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 2.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.5 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 5.4 | GO:0022832 | voltage-gated ion channel activity(GO:0005244) voltage-gated channel activity(GO:0022832) |
| 0.0 | 1.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.8 | GO:0017161 | phosphohistidine phosphatase activity(GO:0008969) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) NADP phosphatase activity(GO:0019178) inositol-4,5-bisphosphate 5-phosphatase activity(GO:0030487) 5-amino-6-(5-phosphoribitylamino)uracil phosphatase activity(GO:0043726) phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) phosphatidylinositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052867) IDP phosphatase activity(GO:1990003) |
| 0.0 | 1.0 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.2 | GO:0070191 | methionine-R-sulfoxide reductase activity(GO:0070191) |
| 0.0 | 1.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 4.2 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 1.9 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.2 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.8 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.8 | GO:0008748 | N-ethylmaleimide reductase activity(GO:0008748) reduced coenzyme F420 dehydrogenase activity(GO:0043738) sulfur oxygenase reductase activity(GO:0043826) malolactic enzyme activity(GO:0043883) epoxyqueuosine reductase activity(GO:0052693) |
| 0.0 | 1.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 1.3 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.5 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.2 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 1.0 | GO:0005216 | ion channel activity(GO:0005216) |
| 0.0 | 2.3 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 1.3 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |