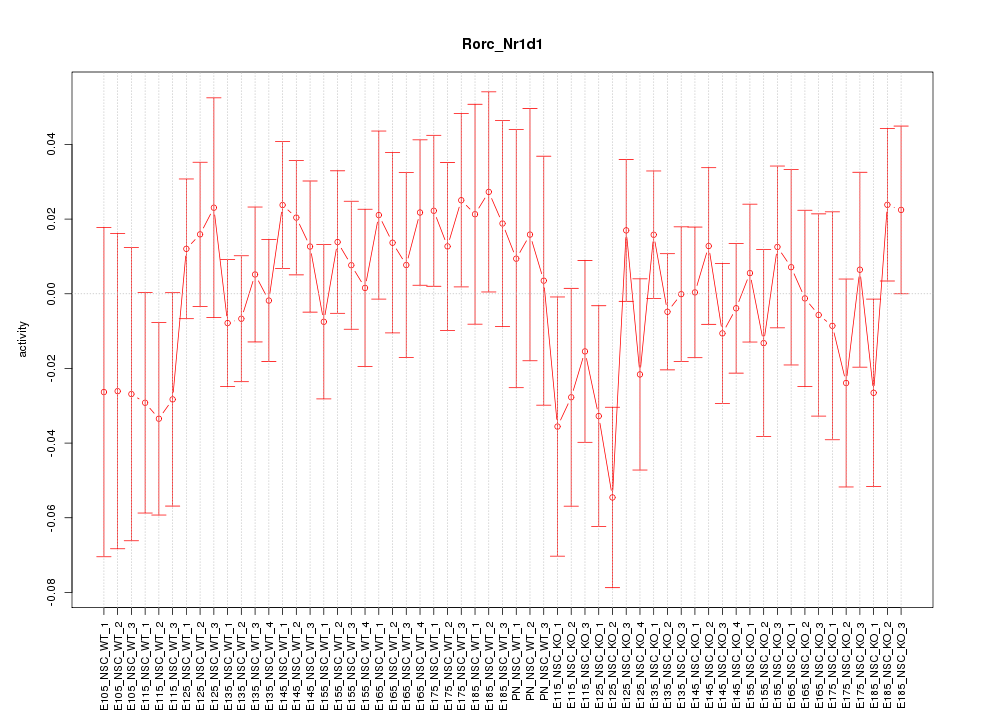
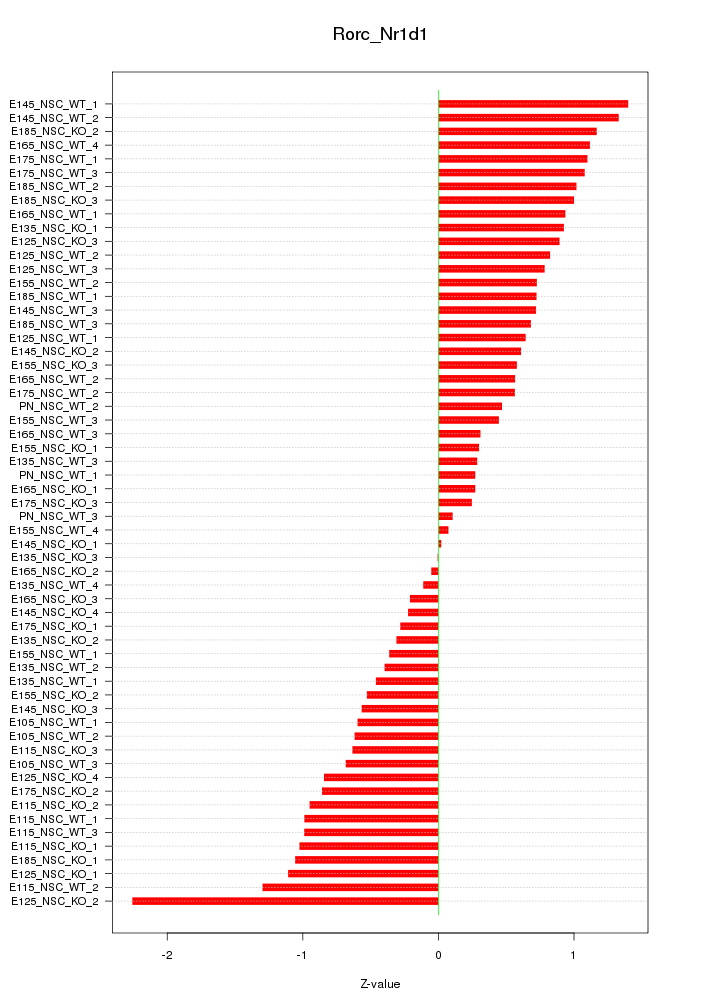
Motif ID: Rorc_Nr1d1
Z-value: 0.789

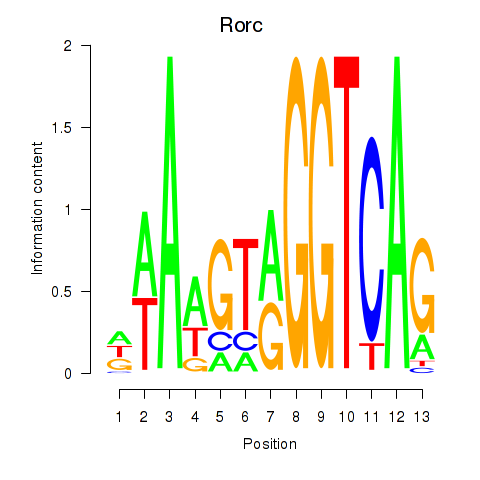
Transcription factors associated with Rorc_Nr1d1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nr1d1 | ENSMUSG00000020889.11 | Nr1d1 |
| Rorc | ENSMUSG00000028150.8 | Rorc |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr1d1 | mm10_v2_chr11_-_98775333_98775354 | 0.44 | 5.7e-04 | Click! |
| Rorc | mm10_v2_chr3_+_94377432_94377451 | 0.19 | 1.4e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.5 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 2.3 | 6.8 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 1.1 | 3.2 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.9 | 2.6 | GO:0060084 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) circadian sleep/wake cycle, REM sleep(GO:0042747) synaptic transmission involved in micturition(GO:0060084) |
| 0.9 | 2.6 | GO:1902336 | neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) positive regulation of retinal ganglion cell axon guidance(GO:1902336) |
| 0.8 | 2.4 | GO:0007210 | serotonin receptor signaling pathway(GO:0007210) |
| 0.5 | 2.8 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.4 | 4.0 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.4 | 1.9 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.3 | 3.1 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.3 | 1.2 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.2 | 0.7 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.2 | 1.7 | GO:0032811 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) negative regulation of epinephrine secretion(GO:0032811) |
| 0.2 | 0.7 | GO:0002554 | serotonin secretion by platelet(GO:0002554) interleukin-3 production(GO:0032632) beta selection(GO:0043366) |
| 0.2 | 1.5 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.2 | 3.2 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.2 | 1.6 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 0.1 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.1 | 2.6 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.1 | 2.1 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.1 | 1.9 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 0.6 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 4.4 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 9.3 | GO:0007601 | visual perception(GO:0007601) |
| 0.1 | 0.8 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.1 | 0.6 | GO:0043313 | regulation of neutrophil degranulation(GO:0043313) |
| 0.1 | 0.4 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.1 | 1.9 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.1 | 0.9 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.1 | 0.6 | GO:0043206 | extracellular fibril organization(GO:0043206) |
| 0.1 | 0.2 | GO:0033860 | regulation of NAD(P)H oxidase activity(GO:0033860) |
| 0.1 | 0.5 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 0.5 | GO:0046036 | GTP biosynthetic process(GO:0006183) CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.3 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 1.5 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 1.1 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.7 | GO:0051974 | negative regulation of telomerase activity(GO:0051974) |
| 0.0 | 0.4 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.5 | GO:0048305 | immunoglobulin secretion(GO:0048305) |
| 0.0 | 1.6 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 3.8 | GO:0007623 | circadian rhythm(GO:0007623) |
| 0.0 | 0.5 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 1.1 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.7 | GO:0046326 | positive regulation of glucose import(GO:0046326) |
| 0.0 | 0.2 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.3 | 1.7 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.2 | 2.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 0.7 | GO:1990879 | CST complex(GO:1990879) |
| 0.2 | 4.0 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.2 | 0.7 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 0.8 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.1 | 0.3 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.9 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 1.6 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 1.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 22.1 | GO:0044297 | cell body(GO:0044297) |
| 0.0 | 1.9 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 2.7 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 2.8 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 1.2 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.5 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 3.2 | GO:0036477 | somatodendritic compartment(GO:0036477) |
| 0.0 | 1.6 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 0.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 9.0 | GO:0005615 | extracellular space(GO:0005615) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.8 | GO:0031752 | D3 dopamine receptor binding(GO:0031750) D5 dopamine receptor binding(GO:0031752) |
| 1.7 | 10.0 | GO:0019841 | retinol binding(GO:0019841) |
| 0.9 | 2.6 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.6 | 3.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.6 | 2.4 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin binding(GO:0051378) serotonin receptor activity(GO:0099589) |
| 0.4 | 1.1 | GO:0004658 | propionyl-CoA carboxylase activity(GO:0004658) |
| 0.3 | 1.9 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.3 | 1.7 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.3 | 2.6 | GO:0004889 | acetylcholine-activated cation-selective channel activity(GO:0004889) |
| 0.3 | 0.8 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.3 | 1.3 | GO:0018121 | imidazoleglycerol-phosphate synthase activity(GO:0000107) NAD(P)-cysteine ADP-ribosyltransferase activity(GO:0018071) NAD(P)-asparagine ADP-ribosyltransferase activity(GO:0018121) NAD(P)-serine ADP-ribosyltransferase activity(GO:0018127) 7-cyano-7-deazaguanine tRNA-ribosyltransferase activity(GO:0043867) purine deoxyribosyltransferase activity(GO:0044102) |
| 0.2 | 1.2 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.2 | 0.6 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.2 | 7.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.9 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 1.5 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 3.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 0.9 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.1 | 10.1 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.1 | 0.7 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.1 | 0.7 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.1 | 1.3 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 1.9 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.7 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 1.9 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 1.7 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 1.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.8 | GO:0050253 | prenylcysteine methylesterase activity(GO:0010296) 1-oxa-2-oxocycloheptane lactonase activity(GO:0018731) sulfolactone hydrolase activity(GO:0018732) butyrolactone hydrolase activity(GO:0018734) endosulfan lactone lactonase activity(GO:0034892) L-ascorbate 6-phosphate lactonase activity(GO:0035460) Ser-tRNA(Thr) hydrolase activity(GO:0043905) Ala-tRNA(Pro) hydrolase activity(GO:0043906) Cys-tRNA(Pro) hydrolase activity(GO:0043907) Ser(Gly)-tRNA(Ala) hydrolase activity(GO:0043908) all-trans-retinyl-palmitate hydrolase, all-trans-retinol forming activity(GO:0047376) retinyl-palmitate esterase activity(GO:0050253) mannosyl-oligosaccharide 1,6-alpha-mannosidase activity(GO:0052767) mannosyl-oligosaccharide 1,3-alpha-mannosidase activity(GO:0052768) methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.0 | 0.5 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 1.9 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.5 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.7 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 2.2 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 1.6 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 1.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.5 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 3.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 1.6 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 3.0 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.1 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |