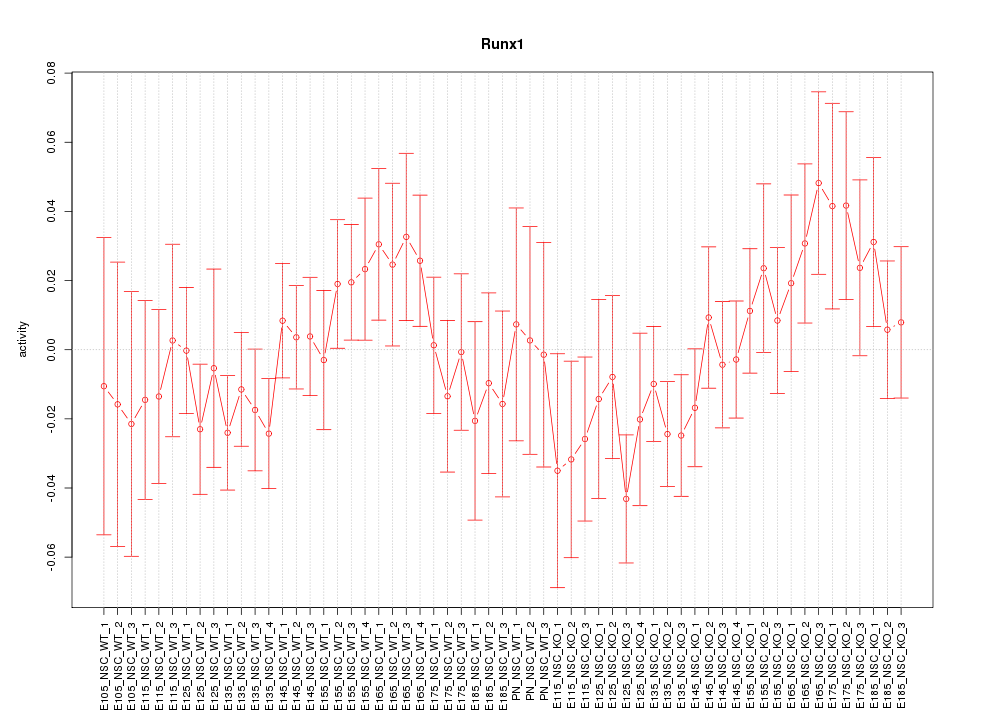
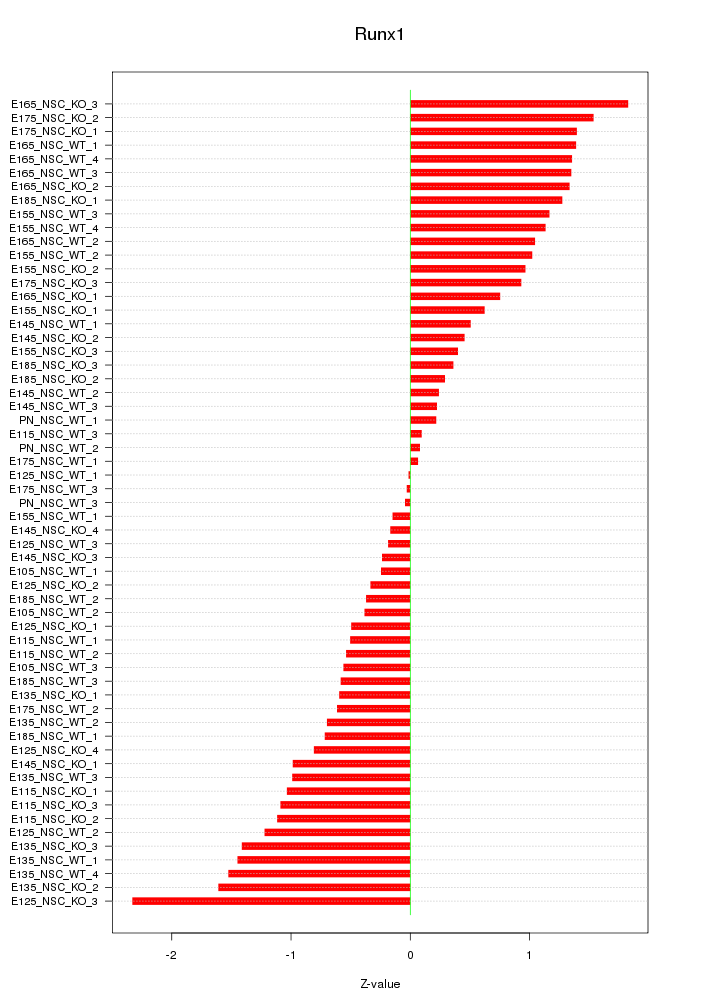
Motif ID: Runx1
Z-value: 0.930
Transcription factors associated with Runx1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Runx1 | ENSMUSG00000022952.10 | Runx1 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 8.5 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 1.1 | 3.3 | GO:2000813 | negative regulation of barbed-end actin filament capping(GO:2000813) |
| 1.0 | 7.0 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.9 | 3.7 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.9 | 4.4 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.8 | 4.0 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.7 | 3.5 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.6 | 3.8 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.6 | 3.8 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.6 | 4.2 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.6 | 3.6 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.5 | 1.4 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.5 | 1.4 | GO:0010752 | signal complex assembly(GO:0007172) regulation of cGMP-mediated signaling(GO:0010752) |
| 0.5 | 2.3 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) positive regulation of receptor binding(GO:1900122) |
| 0.4 | 4.4 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.4 | 2.4 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.4 | 3.3 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.4 | 3.6 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.3 | 1.3 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.3 | 2.0 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.3 | 1.4 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.3 | 1.9 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.2 | 2.3 | GO:0033280 | response to vitamin D(GO:0033280) |
| 0.2 | 2.2 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.2 | 3.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.2 | 7.1 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.2 | 0.7 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.2 | 4.9 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.2 | 2.8 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.2 | 1.7 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 | 2.9 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 2.6 | GO:0048305 | immunoglobulin secretion(GO:0048305) |
| 0.1 | 0.4 | GO:0043313 | regulation of neutrophil degranulation(GO:0043313) |
| 0.1 | 8.6 | GO:0015807 | L-amino acid transport(GO:0015807) |
| 0.1 | 0.6 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.7 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.1 | 5.0 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.1 | 1.1 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 1.2 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.1 | 0.4 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.1 | 11.1 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 1.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 2.5 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.1 | 0.3 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.1 | 7.0 | GO:0030518 | intracellular steroid hormone receptor signaling pathway(GO:0030518) |
| 0.0 | 1.3 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.5 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.4 | GO:0046036 | GTP biosynthetic process(GO:0006183) CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.0 | 0.6 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 2.9 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.6 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.7 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 | 4.7 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 2.2 | GO:0015992 | proton transport(GO:0015992) |
| 0.0 | 0.8 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.3 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.5 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.9 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.4 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.0 | 1.0 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 0.2 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.3 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 1.4 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.3 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.5 | 1.4 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.4 | 3.6 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.4 | 3.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.4 | 4.9 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.3 | 3.3 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.3 | 3.2 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.3 | 1.2 | GO:0031673 | H zone(GO:0031673) |
| 0.2 | 1.2 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.2 | 0.7 | GO:0038045 | large latent transforming growth factor-beta complex(GO:0038045) |
| 0.2 | 4.4 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 3.0 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.1 | 4.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 0.3 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.1 | 4.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 0.4 | GO:1990597 | parallel fiber(GO:1990032) AIP1-IRE1 complex(GO:1990597) |
| 0.1 | 0.7 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.1 | 1.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 4.0 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 1.6 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 3.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 2.9 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 1.5 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 1.5 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 5.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 1.3 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 2.3 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 2.2 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 11.0 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 1.6 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 6.7 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 1.1 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 2.2 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 8.2 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 8.5 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 1.3 | 3.8 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.6 | 7.0 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.6 | 3.7 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.4 | 3.1 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.4 | 1.5 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.3 | 4.0 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.3 | 3.6 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.3 | 3.2 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.3 | 4.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.3 | 3.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.3 | 3.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 5.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.2 | 7.0 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.2 | 9.7 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.2 | 1.3 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.2 | 1.4 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.2 | 0.7 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.2 | 1.4 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 4.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 2.3 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 5.0 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 3.3 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 0.9 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 11.1 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 0.3 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 2.3 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 1.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 2.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 1.2 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 0.7 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 1.9 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 10.3 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 4.5 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.1 | 2.9 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.5 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 1.1 | GO:0035380 | C-3 sterol dehydrogenase (C-4 sterol decarboxylase) activity(GO:0000252) mevaldate reductase activity(GO:0004495) gluconate dehydrogenase activity(GO:0008875) epoxide dehydrogenase activity(GO:0018451) 5-exo-hydroxycamphor dehydrogenase activity(GO:0018452) 2-hydroxytetrahydrofuran dehydrogenase activity(GO:0018453) acetoin dehydrogenase activity(GO:0019152) phenylcoumaran benzylic ether reductase activity(GO:0032442) D-xylose:NADP reductase activity(GO:0032866) L-arabinose:NADP reductase activity(GO:0032867) D-arabinitol dehydrogenase, D-ribulose forming (NADP+) activity(GO:0033709) (R)-(-)-1,2,3,4-tetrahydronaphthol dehydrogenase activity(GO:0034831) 3-hydroxymenthone dehydrogenase activity(GO:0034840) very long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0035380) dihydrotestosterone 17-beta-dehydrogenase activity(GO:0035410) (R)-2-hydroxyisocaproate dehydrogenase activity(GO:0043713) L-arabinose 1-dehydrogenase (NADP+) activity(GO:0044103) L-xylulose reductase (NAD+) activity(GO:0044105) 3-ketoglucose-reductase activity(GO:0048258) D-arabinitol dehydrogenase, D-xylulose forming (NADP+) activity(GO:0052677) |
| 0.0 | 7.1 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 2.6 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 1.0 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 1.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 1.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 1.0 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.8 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 4.1 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |