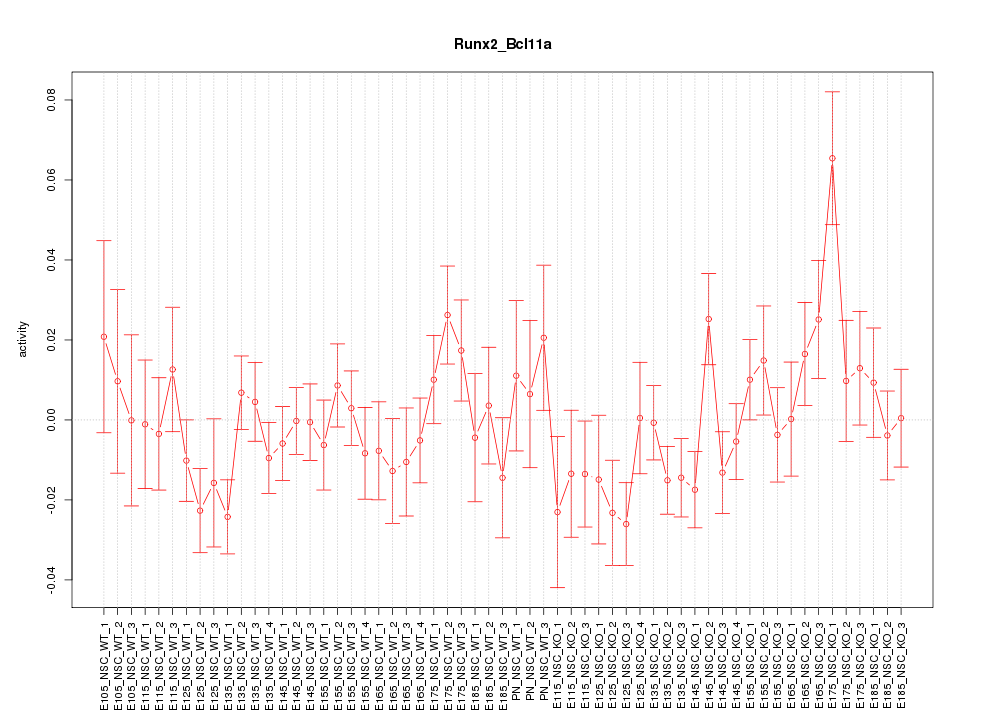
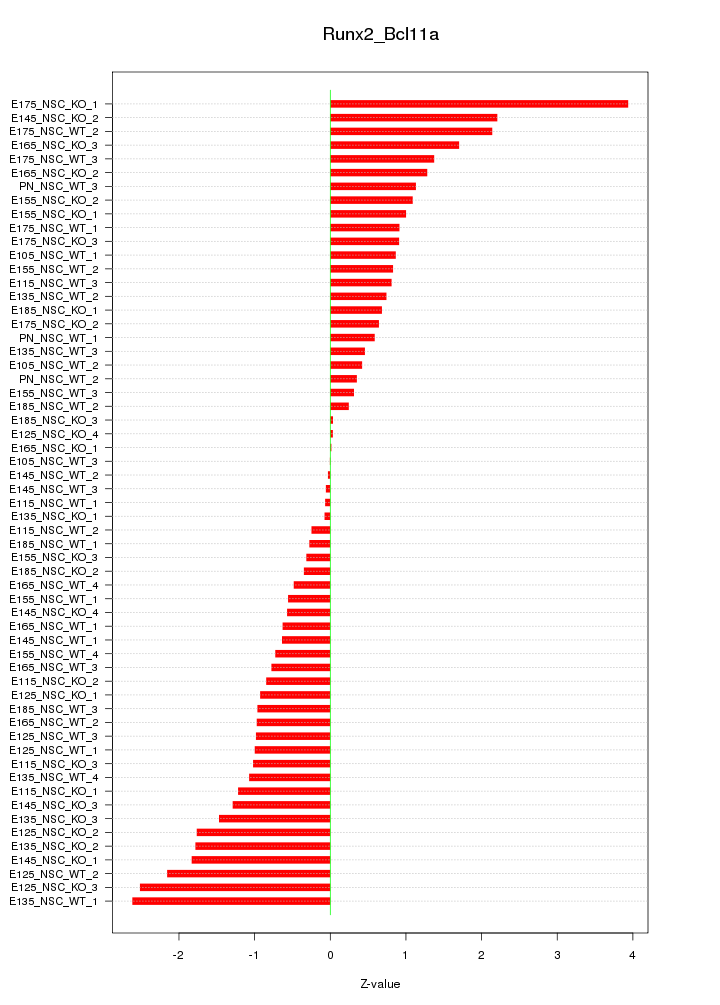
Motif ID: Runx2_Bcl11a
Z-value: 1.198
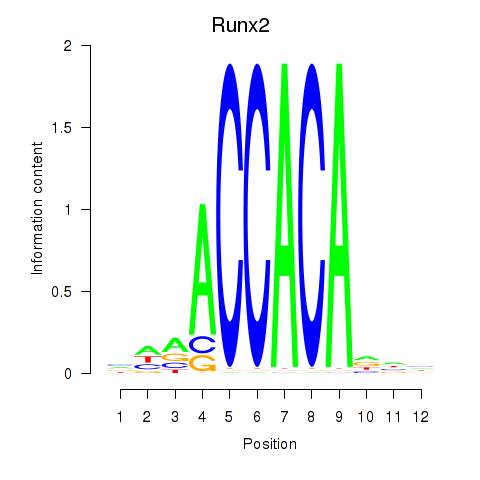
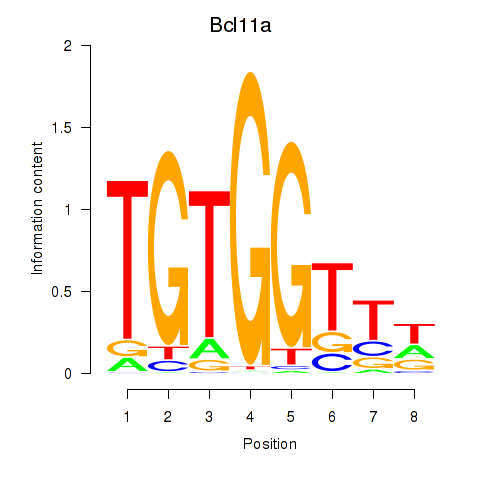
Transcription factors associated with Runx2_Bcl11a:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Bcl11a | ENSMUSG00000000861.9 | Bcl11a |
| Runx2 | ENSMUSG00000039153.10 | Runx2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Bcl11a | mm10_v2_chr11_+_24078173_24078219 | -0.14 | 2.8e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 5.9 | GO:0034162 | toll-like receptor 9 signaling pathway(GO:0034162) |
| 1.8 | 5.5 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 1.8 | 5.5 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 1.3 | 14.0 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 1.3 | 7.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 1.0 | 3.1 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 1.0 | 7.1 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 1.0 | 9.1 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 1.0 | 3.0 | GO:0038095 | positive regulation of mast cell cytokine production(GO:0032765) Fc-epsilon receptor signaling pathway(GO:0038095) |
| 1.0 | 2.9 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 1.0 | 2.9 | GO:0036292 | positive regulation of prostaglandin biosynthetic process(GO:0031394) DNA rewinding(GO:0036292) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.8 | 5.4 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.8 | 3.8 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) negative regulation of cytolysis(GO:0045918) |
| 0.7 | 5.2 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.7 | 2.1 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.6 | 1.9 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.6 | 4.8 | GO:0097460 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) ferrous iron import into cell(GO:0097460) |
| 0.6 | 3.0 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.6 | 1.8 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.6 | 1.8 | GO:0021837 | motogenic signaling involved in postnatal olfactory bulb interneuron migration(GO:0021837) positive regulation of mitotic cell cycle DNA replication(GO:1903465) |
| 0.5 | 2.7 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.5 | 4.3 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.5 | 1.6 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.5 | 2.5 | GO:0010359 | regulation of anion channel activity(GO:0010359) |
| 0.5 | 2.5 | GO:1904721 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.5 | 1.4 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.5 | 3.2 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.5 | 0.9 | GO:2000501 | natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.4 | 4.0 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.4 | 1.3 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.4 | 2.9 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.4 | 2.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.4 | 0.8 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.4 | 1.1 | GO:0002034 | angiotensin mediated vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001998) regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 0.4 | 1.1 | GO:0016115 | terpenoid catabolic process(GO:0016115) |
| 0.4 | 1.8 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.4 | 2.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.4 | 2.1 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.4 | 3.5 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.3 | 0.7 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.3 | 2.4 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.3 | 1.3 | GO:0010813 | neuropeptide catabolic process(GO:0010813) |
| 0.3 | 1.0 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.3 | 1.3 | GO:0021564 | glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 0.3 | 0.6 | GO:0060769 | positive regulation of epithelial cell proliferation involved in prostate gland development(GO:0060769) |
| 0.3 | 1.6 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.3 | 2.2 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.3 | 1.2 | GO:1904177 | regulation of adipose tissue development(GO:1904177) |
| 0.3 | 2.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.3 | 1.2 | GO:0006842 | tricarboxylic acid transport(GO:0006842) succinate transport(GO:0015744) citrate transport(GO:0015746) |
| 0.3 | 1.2 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.3 | 0.9 | GO:0010752 | signal complex assembly(GO:0007172) regulation of cGMP-mediated signaling(GO:0010752) |
| 0.3 | 1.1 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.3 | 0.5 | GO:0001803 | type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 0.3 | 0.8 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.3 | 1.0 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.3 | 1.5 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.2 | 3.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.2 | 0.7 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.2 | 1.0 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.2 | 0.7 | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) |
| 0.2 | 2.5 | GO:0071635 | negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.2 | 0.9 | GO:0019042 | viral latency(GO:0019042) |
| 0.2 | 1.3 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.2 | 2.2 | GO:0033280 | response to vitamin D(GO:0033280) |
| 0.2 | 0.4 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.2 | 2.1 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) |
| 0.2 | 0.8 | GO:1900533 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.2 | 1.0 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.2 | 1.0 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.2 | 1.0 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.2 | 1.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.2 | 0.8 | GO:0010693 | negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.2 | 0.4 | GO:1904529 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.2 | 0.8 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.2 | 1.2 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.2 | 0.6 | GO:0042636 | negative regulation of hair cycle(GO:0042636) negative regulation of hair follicle development(GO:0051799) |
| 0.2 | 0.9 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.2 | 1.2 | GO:0015862 | uridine transport(GO:0015862) |
| 0.2 | 0.5 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.2 | 0.5 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.2 | 0.5 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.2 | 0.5 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.2 | 0.6 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.2 | 0.5 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.2 | 0.5 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.2 | 0.5 | GO:0032829 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) |
| 0.2 | 0.5 | GO:0061526 | acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) positive regulation of synaptic transmission, cholinergic(GO:0032224) acetylcholine secretion(GO:0061526) |
| 0.2 | 2.6 | GO:0017144 | drug metabolic process(GO:0017144) |
| 0.1 | 1.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 0.9 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.1 | 0.4 | GO:2000598 | regulation of cyclin catabolic process(GO:2000598) negative regulation of cyclin catabolic process(GO:2000599) |
| 0.1 | 2.6 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.4 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 | 0.6 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 0.4 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.1 | 0.6 | GO:2000587 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.1 | 2.1 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.5 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.1 | 1.1 | GO:0021612 | facial nerve structural organization(GO:0021612) |
| 0.1 | 1.5 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.1 | 0.3 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 0.1 | 0.4 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.1 | 3.0 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.1 | 0.5 | GO:2001015 | negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.1 | 5.4 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.1 | 0.9 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.1 | 0.3 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.9 | GO:0021894 | cerebral cortex GABAergic interneuron development(GO:0021894) |
| 0.1 | 0.7 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 1.5 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.1 | 2.9 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 3.9 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 0.9 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.1 | 0.9 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.1 | 0.4 | GO:1902514 | regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.1 | 0.6 | GO:2000258 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.1 | 1.3 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 | 0.6 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.6 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.1 | 0.5 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 0.5 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.1 | 0.5 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.1 | 0.8 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 | 0.9 | GO:0021940 | positive regulation of cerebellar granule cell precursor proliferation(GO:0021940) |
| 0.1 | 0.4 | GO:0002827 | positive regulation of T-helper 1 type immune response(GO:0002827) |
| 0.1 | 0.5 | GO:0046502 | uroporphyrinogen III metabolic process(GO:0046502) |
| 0.1 | 0.9 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 0.9 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.1 | 0.7 | GO:0071372 | response to follicle-stimulating hormone(GO:0032354) cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.1 | 1.5 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.1 | 1.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.3 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.1 | 1.2 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.1 | 0.7 | GO:0060055 | angiogenesis involved in wound healing(GO:0060055) |
| 0.1 | 1.4 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 0.4 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.1 | 1.0 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 0.4 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.1 | 1.4 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.1 | 0.3 | GO:0060161 | positive regulation of dopamine receptor signaling pathway(GO:0060161) |
| 0.1 | 0.3 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 1.5 | GO:0010574 | regulation of vascular endothelial growth factor production(GO:0010574) |
| 0.1 | 0.4 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.7 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.1 | 1.6 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.1 | 0.7 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.1 | 0.3 | GO:0051012 | microtubule sliding(GO:0051012) negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.1 | 0.6 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 0.3 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.1 | 0.4 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.1 | 0.7 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.1 | 0.4 | GO:0007031 | peroxisome organization(GO:0007031) |
| 0.1 | 2.3 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.1 | 0.9 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.3 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 0.3 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) saliva secretion(GO:0046541) |
| 0.1 | 0.5 | GO:0043206 | extracellular fibril organization(GO:0043206) |
| 0.1 | 0.4 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 0.3 | GO:0070627 | ferrous iron import(GO:0070627) |
| 0.1 | 0.3 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.1 | 0.4 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) |
| 0.1 | 0.3 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.1 | 0.2 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.1 | 2.8 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.1 | 0.2 | GO:0015669 | gas transport(GO:0015669) carbon dioxide transport(GO:0015670) |
| 0.0 | 0.6 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.0 | 1.8 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.7 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.1 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.0 | 0.3 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.5 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.2 | GO:0060046 | regulation of acrosome reaction(GO:0060046) |
| 0.0 | 0.3 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.3 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.0 | 0.2 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 0.3 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.4 | GO:0006183 | GTP biosynthetic process(GO:0006183) UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.7 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 2.5 | GO:0021766 | hippocampus development(GO:0021766) |
| 0.0 | 0.2 | GO:0002681 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.0 | 0.2 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.2 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.2 | GO:1904417 | regulation of xenophagy(GO:1904415) positive regulation of xenophagy(GO:1904417) |
| 0.0 | 0.4 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.2 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 2.3 | GO:0015807 | L-amino acid transport(GO:0015807) |
| 0.0 | 0.1 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.7 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 1.0 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.2 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.3 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.0 | 0.3 | GO:0036506 | maintenance of unfolded protein(GO:0036506) protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 | 0.3 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.4 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.1 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 1.2 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.1 | GO:0098501 | polynucleotide dephosphorylation(GO:0098501) |
| 0.0 | 0.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 2.2 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.4 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.3 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.0 | 0.2 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.4 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.1 | GO:0010917 | negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.0 | 0.2 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.0 | 0.2 | GO:1903894 | regulation of IRE1-mediated unfolded protein response(GO:1903894) |
| 0.0 | 0.2 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 1.8 | GO:0032091 | negative regulation of protein binding(GO:0032091) |
| 0.0 | 0.6 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.0 | 1.8 | GO:0006818 | hydrogen transport(GO:0006818) |
| 0.0 | 0.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.2 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 1.0 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.1 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.2 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.2 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.1 | GO:2001270 | regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) |
| 0.0 | 0.2 | GO:0051103 | pro-B cell differentiation(GO:0002328) DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.3 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.0 | 1.0 | GO:0019915 | lipid storage(GO:0019915) |
| 0.0 | 0.2 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 1.2 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.6 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.6 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.3 | GO:0051926 | negative regulation of calcium ion transport(GO:0051926) |
| 0.0 | 0.1 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.3 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.9 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.2 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.5 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.5 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.0 | 0.4 | GO:0002690 | positive regulation of leukocyte chemotaxis(GO:0002690) |
| 0.0 | 0.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.5 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.0 | 0.5 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 0.1 | GO:1901032 | negative regulation of response to reactive oxygen species(GO:1901032) negative regulation of hydrogen peroxide-induced cell death(GO:1903206) |
| 0.0 | 0.2 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 0.3 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.2 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.1 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) establishment of protein localization to mitochondrial membrane(GO:0090151) |
| 0.0 | 0.6 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.1 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.0 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.0 | 0.2 | GO:0033006 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.3 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.2 | GO:0090201 | negative regulation of release of cytochrome c from mitochondria(GO:0090201) |
| 0.0 | 0.0 | GO:0001865 | NK T cell differentiation(GO:0001865) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.1 | GO:0061188 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 | 0.0 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.0 | 0.1 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.1 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.3 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.1 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.0 | 0.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 2.1 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.1 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) cellular response to magnesium ion(GO:0071286) regulation of membrane depolarization during action potential(GO:0098902) regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.5 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.0 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.1 | GO:0070571 | negative regulation of neuron projection regeneration(GO:0070571) |
| 0.0 | 0.2 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.3 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.1 | GO:0006956 | complement activation(GO:0006956) |
| 0.0 | 0.1 | GO:0006661 | phosphatidylinositol biosynthetic process(GO:0006661) |
| 0.0 | 0.5 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.1 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.2 | GO:0009268 | response to pH(GO:0009268) |
| 0.0 | 0.0 | GO:0014741 | negative regulation of muscle hypertrophy(GO:0014741) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 18.8 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 1.0 | 3.0 | GO:0038045 | large latent transforming growth factor-beta complex(GO:0038045) |
| 1.0 | 7.8 | GO:0032009 | early phagosome(GO:0032009) |
| 0.7 | 2.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.6 | 4.8 | GO:0097433 | dense body(GO:0097433) |
| 0.6 | 2.8 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.5 | 1.6 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.5 | 3.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.4 | 4.4 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.3 | 1.0 | GO:0044299 | C-fiber(GO:0044299) |
| 0.3 | 0.9 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.3 | 1.8 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.3 | 2.3 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.2 | 0.7 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.2 | 1.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.2 | 17.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.2 | 0.6 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.2 | 3.4 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.2 | 0.6 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.2 | 2.8 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.2 | 1.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.2 | 1.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.2 | 1.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.2 | 1.3 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.2 | 4.0 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 0.5 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.2 | 1.5 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.2 | 0.9 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.2 | 1.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 1.0 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 1.2 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.1 | 0.4 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 3.4 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.1 | 1.4 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 2.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.1 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.1 | 1.6 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 0.3 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.1 | 1.8 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 0.5 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 1.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 1.5 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 2.1 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 0.4 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 0.6 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.1 | 1.0 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 1.7 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.8 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 0.8 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.1 | 0.4 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 1.0 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.6 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 0.6 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 0.5 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 0.7 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 0.3 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.1 | 0.9 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 0.2 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 0.3 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 0.3 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 0.5 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 3.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 0.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 1.1 | GO:0032589 | neuron projection membrane(GO:0032589) |
| 0.0 | 0.9 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.4 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.2 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.4 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 3.8 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.2 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.0 | 10.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 36.0 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.2 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.3 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.6 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 5.6 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.9 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 4.4 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.6 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.4 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.6 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 1.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.6 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.9 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.7 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.5 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.2 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.7 | GO:0099568 | cell cortex(GO:0005938) cytoplasmic region(GO:0099568) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 1.1 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.5 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 8.2 | GO:0019866 | organelle inner membrane(GO:0019866) |
| 0.0 | 3.4 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.5 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.9 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.1 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.3 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 1.0 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.7 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 1.3 | GO:0012506 | vesicle membrane(GO:0012506) |
| 0.0 | 0.2 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 1.0 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.5 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.2 | GO:0000178 | nuclear exosome (RNase complex)(GO:0000176) exosome (RNase complex)(GO:0000178) |
| 0.0 | 1.3 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.1 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 1.1 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.4 | GO:0005901 | caveola(GO:0005901) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 18.8 | GO:0031721 | haptoglobin binding(GO:0031720) hemoglobin alpha binding(GO:0031721) |
| 2.5 | 7.6 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 2.0 | 13.7 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 1.9 | 5.6 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 1.4 | 5.5 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 1.3 | 3.8 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 1.0 | 3.9 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.9 | 3.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.9 | 3.5 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.7 | 2.9 | GO:0070573 | tripeptidyl-peptidase activity(GO:0008240) metallodipeptidase activity(GO:0070573) |
| 0.7 | 2.9 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) phospholipase A2 inhibitor activity(GO:0019834) |
| 0.7 | 5.7 | GO:0035325 | Toll-like receptor binding(GO:0035325) |
| 0.7 | 2.1 | GO:0031755 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.7 | 2.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.7 | 2.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.7 | 4.8 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.7 | 2.7 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.6 | 3.6 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.6 | 1.8 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.6 | 2.3 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.5 | 2.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.5 | 2.5 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.5 | 2.8 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.5 | 10.4 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.4 | 3.5 | GO:0103116 | alpha-D-galactofuranose transporter activity(GO:0103116) |
| 0.4 | 1.3 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.4 | 1.2 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.4 | 1.2 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.4 | 1.2 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.4 | 3.8 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.3 | 1.3 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.3 | 0.3 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.3 | 7.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.3 | 1.0 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.3 | 5.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.3 | 1.3 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.3 | 1.8 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.3 | 5.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.3 | 0.9 | GO:0005118 | sevenless binding(GO:0005118) |
| 0.3 | 0.9 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.3 | 1.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.3 | 1.4 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.3 | 1.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.3 | 0.8 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 0.3 | 2.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.3 | 2.0 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.2 | 1.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.2 | 0.7 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.2 | 0.7 | GO:0015217 | ADP transmembrane transporter activity(GO:0015217) coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.2 | 0.7 | GO:0031752 | D3 dopamine receptor binding(GO:0031750) D5 dopamine receptor binding(GO:0031752) |
| 0.2 | 0.9 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.2 | 0.4 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.2 | 3.2 | GO:0008430 | selenium binding(GO:0008430) |
| 0.2 | 0.9 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.2 | 2.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 1.7 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.2 | 1.0 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 0.6 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) arginine binding(GO:0034618) |
| 0.2 | 1.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.2 | 1.8 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 3.5 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.2 | 1.1 | GO:0019841 | retinal binding(GO:0016918) retinol binding(GO:0019841) |
| 0.2 | 1.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.2 | 0.5 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.2 | 0.9 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.2 | 1.6 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 0.5 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.2 | 1.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.2 | 1.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 1.5 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.1 | 1.5 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.1 | 1.8 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 1.7 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.5 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.4 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.1 | 4.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.4 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 2.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 2.8 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 1.3 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.1 | 0.9 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 0.7 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 1.5 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 3.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 0.6 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) |
| 0.1 | 4.3 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.1 | 1.2 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.1 | 1.6 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 1.0 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.3 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) |
| 0.1 | 1.4 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 0.5 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 0.4 | GO:0016936 | beta-galactosidase activity(GO:0004565) galactoside binding(GO:0016936) |
| 0.1 | 0.5 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.1 | 0.9 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.3 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 0.9 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 0.6 | GO:0070699 | inhibin binding(GO:0034711) type II activin receptor binding(GO:0070699) |
| 0.1 | 0.6 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.1 | 0.6 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.1 | 0.6 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 1.2 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 0.4 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.1 | 0.3 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.1 | 3.3 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 1.0 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.8 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 2.9 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 1.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.4 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.4 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 0.7 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 0.5 | GO:0045703 | pinocarveol dehydrogenase activity(GO:0018446) chloral hydrate dehydrogenase activity(GO:0018447) hydroxymethylmethylsilanediol oxidase activity(GO:0018448) 1-phenylethanol dehydrogenase activity(GO:0018449) myrtenol dehydrogenase activity(GO:0018450) cis-1,2-dihydroxy-1,2-dihydro-8-carboxynaphthalene dehydrogenase activity(GO:0034522) 3-hydroxy-4-methyloctanoyl-CoA dehydrogenase activity(GO:0034582) 2-hydroxy-4-isopropenylcyclohexane-1-carboxyl-CoA dehydrogenase activity(GO:0034778) cis-9,10-dihydroanthracene-9,10-diol dehydrogenase activity(GO:0034817) citronellol dehydrogenase activity(GO:0034821) naphthyl-2-hydroxymethyl-succinyl-CoA dehydrogenase activity(GO:0034847) 2,4,4-trimethyl-1-pentanol dehydrogenase activity(GO:0034863) 2,4,4-trimethyl-3-hydroxypentanoyl-CoA dehydrogenase activity(GO:0034868) 1-hydroxy-4,4-dimethylpentan-3-one dehydrogenase activity(GO:0034871) endosulfan diol dehydrogenase activity(GO:0034891) endosulfan hydroxyether dehydrogenase activity(GO:0034901) 3-hydroxy-2-methylhexanoyl-CoA dehydrogenase activity(GO:0034918) 3-hydroxy-2,6-dimethyl-5-methylene-heptanoyl-CoA dehydrogenase activity(GO:0034944) versicolorin reductase activity(GO:0042469) ketoreductase activity(GO:0045703) |
| 0.1 | 3.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.2 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 0.7 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.1 | 0.5 | GO:0031802 | G-protein coupled adenosine receptor activity(GO:0001609) type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.1 | 0.9 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 0.2 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.1 | 5.1 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.1 | 0.7 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 0.4 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 0.8 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 0.3 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.3 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.1 | 0.5 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.3 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 0.1 | GO:0004637 | phosphoribosylamine-glycine ligase activity(GO:0004637) |
| 0.1 | 0.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.8 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.6 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 2.1 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 1.0 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 1.2 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0097100 | oxidized purine nucleobase lesion DNA N-glycosylase activity(GO:0008534) supercoiled DNA binding(GO:0097100) |
| 0.0 | 0.2 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.1 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.0 | 3.3 | GO:0043492 | ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 0.7 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.7 | GO:0034946 | 2-oxoglutaryl-CoA thioesterase activity(GO:0034843) 2,4,4-trimethyl-3-oxopentanoyl-CoA thioesterase activity(GO:0034869) 3-isopropylbut-3-enoyl-CoA thioesterase activity(GO:0034946) glutaryl-CoA hydrolase activity(GO:0044466) |
| 0.0 | 1.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 1.8 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 1.2 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.4 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.7 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.3 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.4 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.4 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.6 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.2 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.6 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.3 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.0 | 0.2 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.3 | GO:0035004 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) phosphatidylinositol 3-kinase activity(GO:0035004) |
| 0.0 | 1.9 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.3 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 3.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.6 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.9 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.7 | GO:0034945 | dihydrolipoamide branched chain acyltransferase activity(GO:0004147) palmitoleoyl [acyl-carrier-protein]-dependent acyltransferase activity(GO:0008951) serine O-acyltransferase activity(GO:0016412) O-succinyltransferase activity(GO:0016750) sinapoyltransferase activity(GO:0016752) O-sinapoyltransferase activity(GO:0016753) peptidyl-lysine N6-myristoyltransferase activity(GO:0018030) peptidyl-lysine N6-palmitoyltransferase activity(GO:0018031) benzoyl acetate-CoA thiolase activity(GO:0018711) 3-hydroxybutyryl-CoA thiolase activity(GO:0018712) 3-ketopimelyl-CoA thiolase activity(GO:0018713) N-palmitoyltransferase activity(GO:0019105) acyl-CoA N-acyltransferase activity(GO:0019186) protein-cysteine S-myristoyltransferase activity(GO:0019705) glucosaminyl-phosphotidylinositol O-acyltransferase activity(GO:0032216) ergosterol O-acyltransferase activity(GO:0034737) lanosterol O-acyltransferase activity(GO:0034738) naphthyl-2-oxomethyl-succinyl-CoA succinyl transferase activity(GO:0034848) 2,4,4-trimethyl-3-oxopentanoyl-CoA 2-C-propanoyl transferase activity(GO:0034851) 2-methylhexanoyl-CoA C-acetyltransferase activity(GO:0034915) butyryl-CoA 2-C-propionyltransferase activity(GO:0034919) 2,6-dimethyl-5-methylene-3-oxo-heptanoyl-CoA C-acetyltransferase activity(GO:0034945) L-2-aminoadipate N-acetyltransferase activity(GO:0043741) keto acid formate lyase activity(GO:0043806) azetidine-2-carboxylic acid acetyltransferase activity(GO:0046941) peptidyl-lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0052858) acetyl-CoA:L-lysine N6-acetyltransferase(GO:0090595) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.4 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 1.3 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.7 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.7 | GO:0008748 | N-ethylmaleimide reductase activity(GO:0008748) reduced coenzyme F420 dehydrogenase activity(GO:0043738) sulfur oxygenase reductase activity(GO:0043826) malolactic enzyme activity(GO:0043883) epoxyqueuosine reductase activity(GO:0052693) |
| 0.0 | 0.7 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.1 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.1 | GO:0046403 | polynucleotide 3'-phosphatase activity(GO:0046403) |
| 0.0 | 0.4 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.1 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.2 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.9 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 1.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.0 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 1.5 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.1 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.2 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.0 | 0.0 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.4 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.1 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.7 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.2 | GO:0004707 | MAP kinase activity(GO:0004707) |