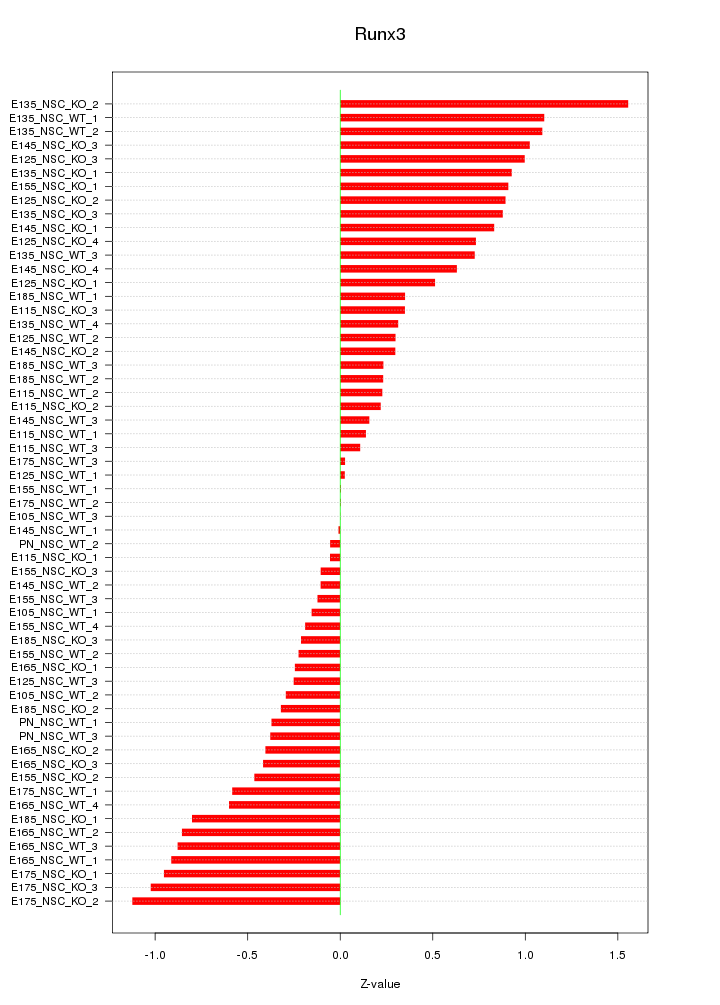
Motif ID: Runx3
Z-value: 0.605

Transcription factors associated with Runx3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Runx3 | ENSMUSG00000070691.4 | Runx3 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0003195 | tricuspid valve development(GO:0003175) tricuspid valve morphogenesis(GO:0003186) tricuspid valve formation(GO:0003195) right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.5 | 9.2 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.5 | 4.8 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.4 | 1.6 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.4 | 1.2 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.4 | 1.1 | GO:0021893 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 0.2 | 0.9 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.2 | 0.5 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.1 | 0.7 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.1 | 0.9 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.1 | 0.9 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) regulation of wound healing, spreading of epidermal cells(GO:1903689) |
| 0.1 | 5.1 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.1 | 0.4 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.1 | 2.0 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.1 | 1.5 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.1 | 0.8 | GO:0046036 | CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.1 | 1.5 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 0.4 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 3.2 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.1 | 0.1 | GO:0014891 | striated muscle atrophy(GO:0014891) |
| 0.1 | 0.3 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 0.4 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.8 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.7 | GO:0034724 | DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 | 0.5 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.0 | 0.6 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.6 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.5 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.9 | GO:0043330 | response to exogenous dsRNA(GO:0043330) |
| 0.0 | 0.7 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.7 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.3 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.5 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 1.2 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.8 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.4 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.5 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 1.3 | GO:0007127 | meiosis I(GO:0007127) |
| 0.0 | 0.4 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.2 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 | 1.2 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 5.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.5 | 3.2 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.4 | 1.2 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 0.9 | GO:0030677 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.1 | 9.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 0.9 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 1.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.3 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.1 | 0.4 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.7 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.9 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 1.5 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 2.5 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.5 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.7 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.3 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 5.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.4 | 4.8 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.2 | 0.8 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.2 | 0.5 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.2 | 0.6 | GO:0098634 | protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.4 | GO:0048045 | 4-hydroxybenzoate octaprenyltransferase activity(GO:0008412) protoheme IX farnesyltransferase activity(GO:0008495) (S)-2,3-di-O-geranylgeranylglyceryl phosphate synthase activity(GO:0043888) cadaverine aminopropyltransferase activity(GO:0043918) agmatine aminopropyltransferase activity(GO:0043919) 1,4-dihydroxy-2-naphthoate octaprenyltransferase activity(GO:0046428) trans-pentaprenyltranstransferase activity(GO:0048045) ATP dimethylallyltransferase activity(GO:0052622) ADP dimethylallyltransferase activity(GO:0052623) |
| 0.1 | 0.9 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 0.5 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.1 | 0.5 | GO:0043842 | Kdo transferase activity(GO:0043842) |
| 0.1 | 0.3 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.1 | 0.9 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.1 | 0.9 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 0.9 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.7 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.4 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 1.2 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 2.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 9.8 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.8 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.3 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 1.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.5 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.6 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 0.2 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.4 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.4 | GO:0044653 | trehalase activity(GO:0015927) dextrin alpha-glucosidase activity(GO:0044653) starch alpha-glucosidase activity(GO:0044654) beta-glucanase activity(GO:0052736) beta-6-sulfate-N-acetylglucosaminidase activity(GO:0052769) glucan endo-1,4-beta-glucosidase activity(GO:0052859) |
| 0.0 | 0.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.7 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 1.1 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 2.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.9 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.7 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.7 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |