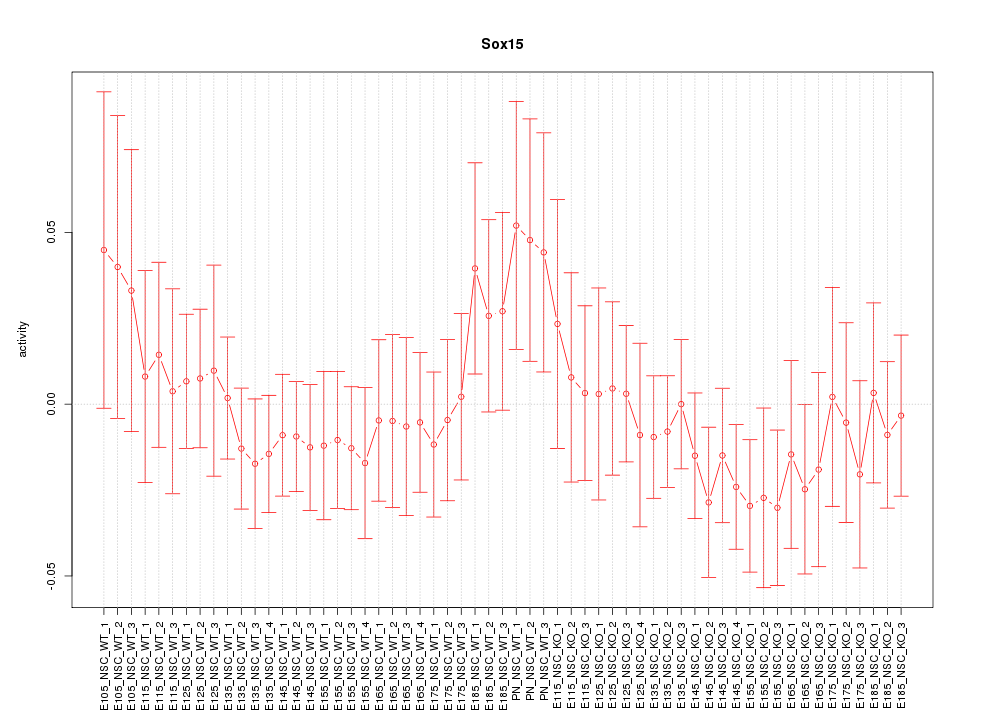
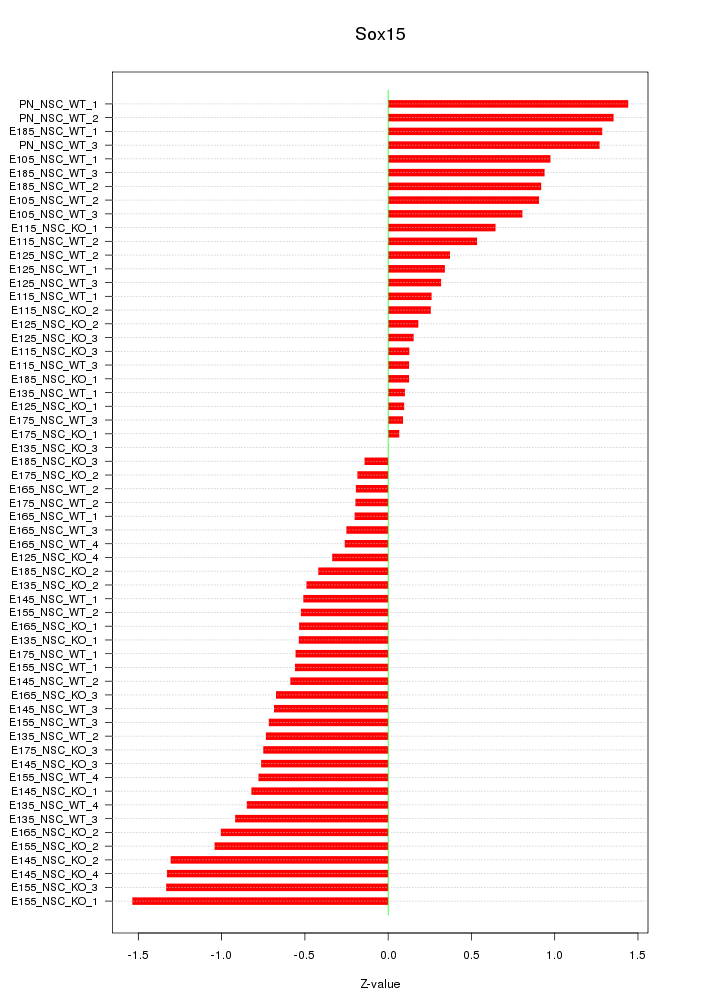
Motif ID: Sox15
Z-value: 0.727
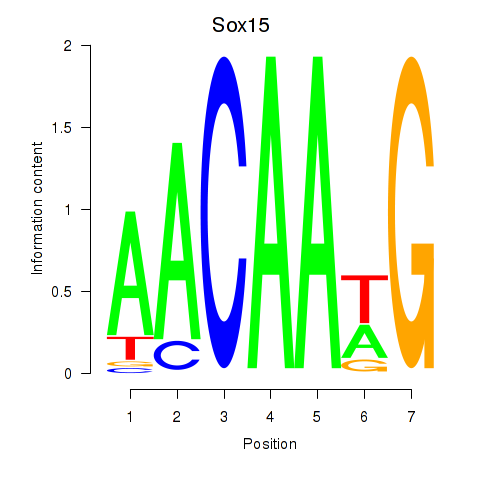
Transcription factors associated with Sox15:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Sox15 | ENSMUSG00000041287.5 | Sox15 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.2 | GO:0050904 | diapedesis(GO:0050904) |
| 1.4 | 5.8 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 1.1 | 8.0 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 1.1 | 4.5 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 1.1 | 4.3 | GO:1904192 | prostate gland stromal morphogenesis(GO:0060741) cholangiocyte apoptotic process(GO:1902488) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 1.0 | 5.8 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.9 | 2.7 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) odontoblast differentiation(GO:0071895) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.9 | 6.2 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.8 | 2.4 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.8 | 2.3 | GO:0042320 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) circadian sleep/wake cycle, REM sleep(GO:0042747) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.8 | 3.1 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.8 | 3.8 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.7 | 3.7 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.7 | 2.2 | GO:0097274 | urea homeostasis(GO:0097274) |
| 0.7 | 5.6 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.6 | 3.2 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.6 | 1.7 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.5 | 2.6 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.4 | 4.3 | GO:0009186 | deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.4 | 1.7 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.4 | 2.8 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.4 | 1.5 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.4 | 1.4 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.3 | 1.0 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.3 | 4.8 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.3 | 5.4 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.2 | 7.7 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-dependent nucleosome organization(GO:0034723) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.2 | 1.2 | GO:2000325 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.2 | 1.3 | GO:0051461 | regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.2 | 0.6 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.2 | 1.5 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.2 | 1.5 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.2 | 5.1 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.2 | 3.1 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.2 | 1.4 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 2.4 | GO:0030903 | notochord development(GO:0030903) |
| 0.1 | 0.7 | GO:0051036 | regulation of endosome size(GO:0051036) |
| 0.1 | 1.3 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 1.8 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 0.9 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 0.7 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 1.8 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 0.4 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.1 | 1.3 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 1.0 | GO:0006337 | nucleosome disassembly(GO:0006337) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 0.4 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 1.7 | GO:0009409 | response to cold(GO:0009409) |
| 0.1 | 1.4 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.1 | 4.2 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.0 | 3.3 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.4 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 1.3 | GO:2000772 | regulation of cellular senescence(GO:2000772) |
| 0.0 | 2.1 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.0 | 0.2 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.0 | 5.5 | GO:0042552 | myelination(GO:0042552) |
| 0.0 | 0.8 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.3 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.8 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 2.6 | GO:0030168 | platelet activation(GO:0030168) |
| 0.0 | 1.7 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.6 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.9 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 2.8 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 1.9 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 6.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 1.2 | 6.2 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 1.2 | 3.5 | GO:0042585 | germinal vesicle(GO:0042585) |
| 1.1 | 4.3 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.9 | 4.3 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.8 | 8.3 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.7 | 5.5 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.5 | 2.6 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.4 | 4.8 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.3 | 1.3 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.2 | 5.1 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.2 | 1.0 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.2 | 1.8 | GO:0031105 | septin complex(GO:0031105) |
| 0.1 | 2.4 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.1 | 1.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 1.7 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 0.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 0.3 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.1 | 2.8 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 1.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.7 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 3.3 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 1.2 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 1.0 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 3.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 8.1 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 3.2 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 1.8 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 2.2 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 3.3 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 3.6 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 9.9 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.4 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 1.5 | GO:0005774 | vacuolar membrane(GO:0005774) |
| 0.0 | 4.5 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 1.1 | 4.5 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.9 | 4.3 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.8 | 2.4 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.7 | 8.0 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.5 | 2.7 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.4 | 6.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.4 | 3.8 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
| 0.4 | 1.5 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.4 | 3.7 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.3 | 2.6 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.3 | 1.3 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.3 | 5.6 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.2 | 1.7 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 4.9 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.2 | 1.4 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.2 | 1.4 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.2 | 1.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 4.3 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.2 | 3.2 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.2 | 0.6 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 2.2 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.1 | 1.7 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 2.3 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 1.5 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.1 | 1.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 1.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 3.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 1.8 | GO:0034945 | dihydrolipoamide branched chain acyltransferase activity(GO:0004147) palmitoleoyl [acyl-carrier-protein]-dependent acyltransferase activity(GO:0008951) serine O-acyltransferase activity(GO:0016412) O-succinyltransferase activity(GO:0016750) sinapoyltransferase activity(GO:0016752) O-sinapoyltransferase activity(GO:0016753) peptidyl-lysine N6-myristoyltransferase activity(GO:0018030) peptidyl-lysine N6-palmitoyltransferase activity(GO:0018031) benzoyl acetate-CoA thiolase activity(GO:0018711) 3-hydroxybutyryl-CoA thiolase activity(GO:0018712) 3-ketopimelyl-CoA thiolase activity(GO:0018713) N-palmitoyltransferase activity(GO:0019105) acyl-CoA N-acyltransferase activity(GO:0019186) protein-cysteine S-myristoyltransferase activity(GO:0019705) glucosaminyl-phosphotidylinositol O-acyltransferase activity(GO:0032216) ergosterol O-acyltransferase activity(GO:0034737) lanosterol O-acyltransferase activity(GO:0034738) naphthyl-2-oxomethyl-succinyl-CoA succinyl transferase activity(GO:0034848) 2,4,4-trimethyl-3-oxopentanoyl-CoA 2-C-propanoyl transferase activity(GO:0034851) 2-methylhexanoyl-CoA C-acetyltransferase activity(GO:0034915) butyryl-CoA 2-C-propionyltransferase activity(GO:0034919) 2,6-dimethyl-5-methylene-3-oxo-heptanoyl-CoA C-acetyltransferase activity(GO:0034945) L-2-aminoadipate N-acetyltransferase activity(GO:0043741) keto acid formate lyase activity(GO:0043806) azetidine-2-carboxylic acid acetyltransferase activity(GO:0046941) peptidyl-lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0052858) acetyl-CoA:L-lysine N6-acetyltransferase(GO:0090595) |
| 0.1 | 2.1 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 0.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 3.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 1.0 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 2.9 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 1.4 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 6.1 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 13.4 | GO:0000987 | core promoter proximal region sequence-specific DNA binding(GO:0000987) |
| 0.0 | 0.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.4 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.3 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 1.9 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.4 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 2.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.9 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 1.6 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.5 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 1.0 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |