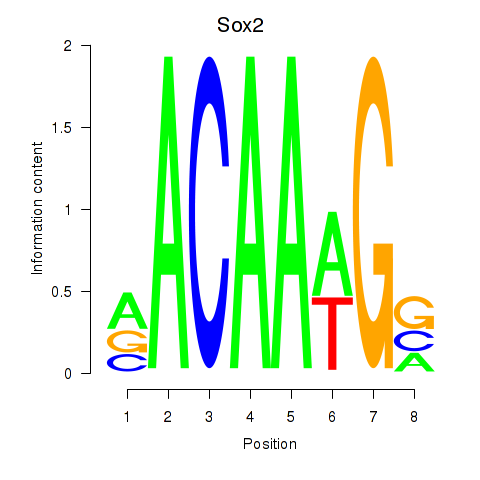
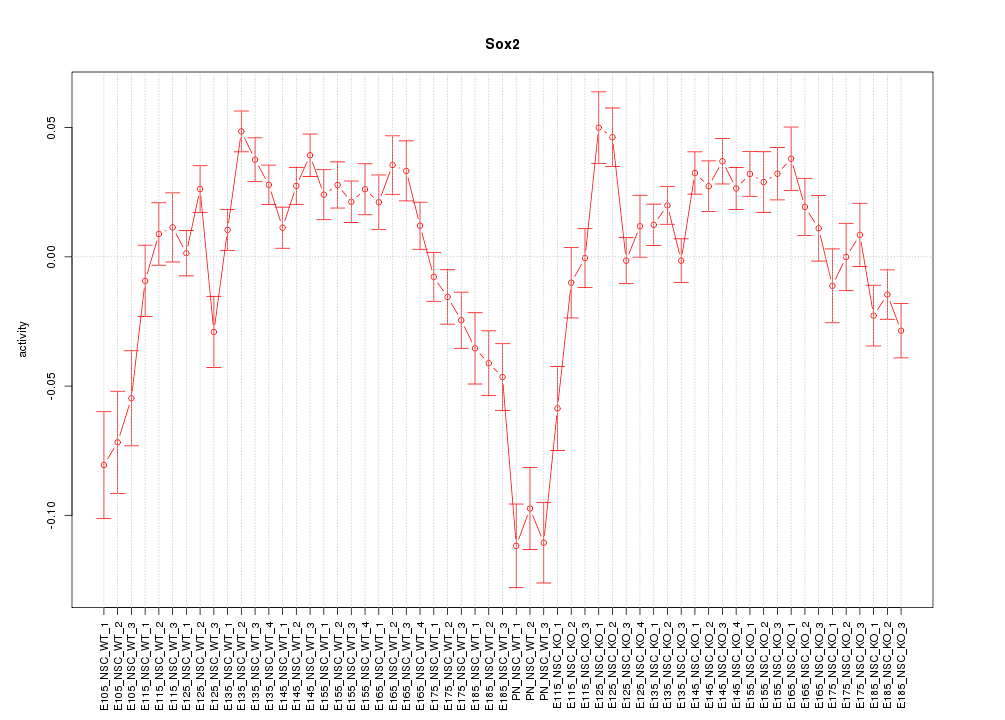
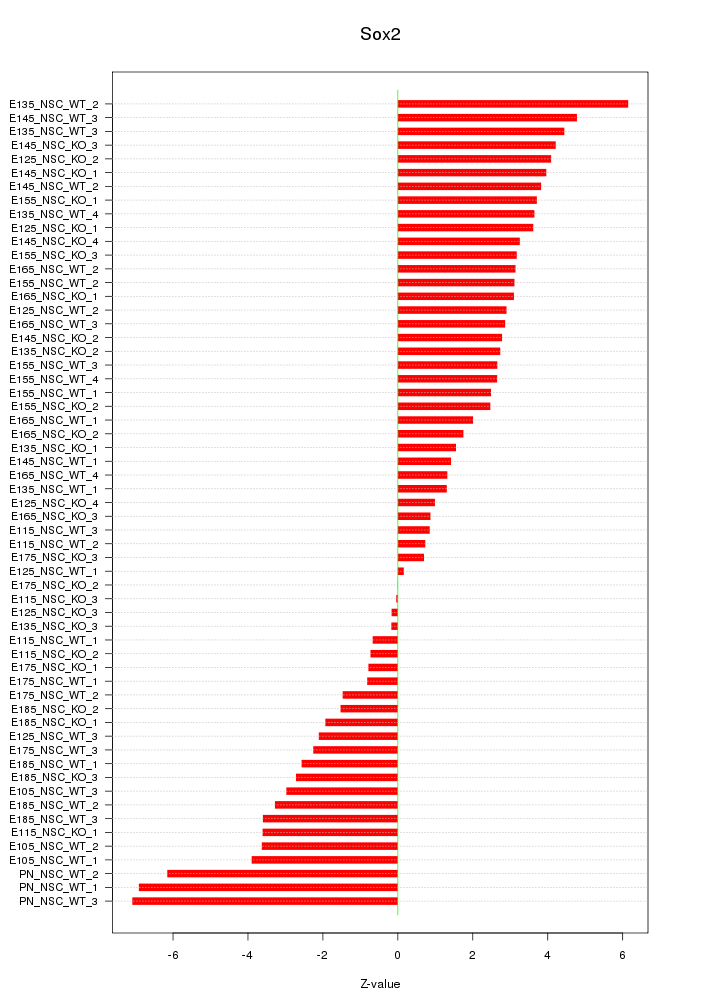
| 9.8 |
29.4 |
GO:2000297 |
negative regulation of synapse maturation(GO:2000297) |
| 6.0 |
17.9 |
GO:0071929 |
alpha-tubulin acetylation(GO:0071929) |
| 5.8 |
17.4 |
GO:2000344 |
positive regulation of acrosome reaction(GO:2000344) |
| 5.5 |
16.5 |
GO:0030827 |
negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 5.2 |
25.9 |
GO:0031022 |
nuclear migration along microfilament(GO:0031022) |
| 4.9 |
14.6 |
GO:0042560 |
10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 4.6 |
23.2 |
GO:0046684 |
response to pyrethroid(GO:0046684) |
| 4.6 |
104.7 |
GO:0021819 |
layer formation in cerebral cortex(GO:0021819) |
| 4.2 |
12.5 |
GO:0003345 |
proepicardium cell migration involved in pericardium morphogenesis(GO:0003345) |
| 3.9 |
11.8 |
GO:2000620 |
positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 3.8 |
11.5 |
GO:0002302 |
CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 3.8 |
11.4 |
GO:0003245 |
cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 3.7 |
40.8 |
GO:0010650 |
positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 3.7 |
18.4 |
GO:2001025 |
positive regulation of response to drug(GO:2001025) |
| 3.6 |
14.4 |
GO:0060316 |
positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 3.5 |
17.6 |
GO:0043314 |
negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 3.4 |
17.1 |
GO:0035984 |
response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 3.3 |
10.0 |
GO:0090327 |
negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 3.3 |
9.9 |
GO:0017085 |
response to insecticide(GO:0017085) |
| 3.3 |
52.4 |
GO:1900454 |
positive regulation of long term synaptic depression(GO:1900454) |
| 3.3 |
39.2 |
GO:0031274 |
positive regulation of pseudopodium assembly(GO:0031274) |
| 3.2 |
13.0 |
GO:1904529 |
regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 3.2 |
9.7 |
GO:0021649 |
vestibulocochlear nerve morphogenesis(GO:0021648) vestibulocochlear nerve structural organization(GO:0021649) ganglion morphogenesis(GO:0061552) dorsal root ganglion morphogenesis(GO:1904835) dorsal root ganglion development(GO:1990791) |
| 3.2 |
16.1 |
GO:1903715 |
regulation of aerobic respiration(GO:1903715) |
| 3.2 |
9.6 |
GO:0002842 |
T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) positive regulation of T cell mediated immune response to tumor cell(GO:0002842) positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 3.1 |
18.7 |
GO:0006172 |
ADP biosynthetic process(GO:0006172) |
| 3.0 |
11.9 |
GO:0046898 |
response to cycloheximide(GO:0046898) |
| 2.9 |
11.7 |
GO:0036072 |
intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 2.8 |
11.3 |
GO:0086042 |
cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) |
| 2.8 |
8.5 |
GO:0070428 |
regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 2.7 |
10.9 |
GO:0001922 |
B-1 B cell homeostasis(GO:0001922) |
| 2.6 |
7.9 |
GO:0007403 |
glial cell fate determination(GO:0007403) |
| 2.6 |
15.6 |
GO:0061549 |
sympathetic ganglion development(GO:0061549) |
| 2.5 |
7.6 |
GO:0021933 |
radial glia guided migration of cerebellar granule cell(GO:0021933) |
| 2.5 |
7.6 |
GO:0070649 |
formin-nucleated actin cable assembly(GO:0070649) |
| 2.5 |
12.6 |
GO:0030259 |
lipid glycosylation(GO:0030259) |
| 2.5 |
12.4 |
GO:0036515 |
serotonergic neuron axon guidance(GO:0036515) |
| 2.5 |
7.4 |
GO:0060478 |
acrosomal vesicle exocytosis(GO:0060478) |
| 2.4 |
19.6 |
GO:0061368 |
behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 2.4 |
12.0 |
GO:0060158 |
phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 2.4 |
9.5 |
GO:0097393 |
post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 2.4 |
7.1 |
GO:0038095 |
positive regulation of mast cell cytokine production(GO:0032765) Fc-epsilon receptor signaling pathway(GO:0038095) |
| 2.4 |
9.4 |
GO:0033326 |
cerebrospinal fluid secretion(GO:0033326) |
| 2.3 |
20.7 |
GO:0035507 |
regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 2.3 |
36.7 |
GO:0035414 |
negative regulation of catenin import into nucleus(GO:0035414) |
| 2.3 |
4.6 |
GO:2000017 |
positive regulation of determination of dorsal identity(GO:2000017) |
| 2.3 |
4.5 |
GO:0021886 |
hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 2.3 |
9.1 |
GO:1900533 |
medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 2.3 |
9.0 |
GO:1900740 |
regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 2.2 |
6.5 |
GO:0014877 |
response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 2.1 |
8.3 |
GO:1990743 |
protein sialylation(GO:1990743) |
| 2.1 |
8.3 |
GO:0060729 |
intestinal epithelial structure maintenance(GO:0060729) |
| 1.9 |
13.3 |
GO:0034047 |
regulation of protein phosphatase type 2A activity(GO:0034047) |
| 1.8 |
7.4 |
GO:0034214 |
protein hexamerization(GO:0034214) |
| 1.8 |
7.2 |
GO:0061428 |
embryonic heart tube left/right pattern formation(GO:0060971) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 1.7 |
5.2 |
GO:0060392 |
negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 1.7 |
17.3 |
GO:0007379 |
segment specification(GO:0007379) |
| 1.6 |
6.5 |
GO:0010499 |
proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 1.6 |
6.4 |
GO:2000795 |
negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 1.6 |
9.4 |
GO:0061343 |
cell adhesion involved in heart morphogenesis(GO:0061343) |
| 1.6 |
4.7 |
GO:0009256 |
10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 1.5 |
6.1 |
GO:0000415 |
negative regulation of histone H3-K36 methylation(GO:0000415) |
| 1.5 |
4.5 |
GO:1902460 |
regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 1.5 |
4.4 |
GO:0036092 |
phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 1.5 |
16.0 |
GO:0071225 |
cellular response to muramyl dipeptide(GO:0071225) |
| 1.4 |
4.3 |
GO:2000096 |
regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 1.4 |
4.3 |
GO:0015712 |
hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 1.4 |
7.1 |
GO:0035262 |
gonad morphogenesis(GO:0035262) |
| 1.4 |
4.3 |
GO:0061588 |
calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 1.4 |
26.5 |
GO:0086036 |
regulation of cardiac muscle cell membrane potential(GO:0086036) |
| 1.4 |
19.1 |
GO:0050901 |
leukocyte tethering or rolling(GO:0050901) |
| 1.3 |
9.0 |
GO:1903300 |
negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 1.3 |
5.1 |
GO:0007412 |
axon target recognition(GO:0007412) |
| 1.3 |
7.6 |
GO:0060164 |
regulation of timing of neuron differentiation(GO:0060164) |
| 1.2 |
2.5 |
GO:0033088 |
negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 1.2 |
7.2 |
GO:0000160 |
phosphorelay signal transduction system(GO:0000160) |
| 1.2 |
10.8 |
GO:0048505 |
regulation of timing of cell differentiation(GO:0048505) |
| 1.2 |
15.2 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 1.1 |
8.9 |
GO:0035881 |
amacrine cell differentiation(GO:0035881) |
| 1.1 |
4.5 |
GO:0051562 |
negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 1.1 |
14.3 |
GO:0043252 |
sodium-independent organic anion transport(GO:0043252) |
| 1.1 |
2.2 |
GO:0051572 |
negative regulation of histone H3-K4 methylation(GO:0051572) |
| 1.1 |
21.5 |
GO:0006054 |
N-acetylneuraminate metabolic process(GO:0006054) |
| 1.1 |
3.2 |
GO:2000744 |
anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 1.1 |
8.4 |
GO:1900107 |
regulation of nodal signaling pathway(GO:1900107) |
| 1.0 |
4.1 |
GO:2000427 |
apolipoprotein A-I-mediated signaling pathway(GO:0038027) regulation of apoptotic cell clearance(GO:2000425) positive regulation of apoptotic cell clearance(GO:2000427) |
| 1.0 |
6.1 |
GO:0031584 |
activation of phospholipase D activity(GO:0031584) |
| 1.0 |
6.0 |
GO:1902950 |
regulation of dendritic spine maintenance(GO:1902950) positive regulation of dendritic spine maintenance(GO:1902952) |
| 1.0 |
4.0 |
GO:0043973 |
histone H3-K4 acetylation(GO:0043973) |
| 1.0 |
3.8 |
GO:0044861 |
protein transport into plasma membrane raft(GO:0044861) |
| 0.9 |
7.6 |
GO:0032625 |
interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.9 |
16.1 |
GO:0007194 |
negative regulation of adenylate cyclase activity(GO:0007194) negative regulation of cyclase activity(GO:0031280) |
| 0.9 |
6.6 |
GO:2000394 |
positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.9 |
31.9 |
GO:0043551 |
regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.9 |
3.7 |
GO:0072697 |
protein localization to cell cortex(GO:0072697) |
| 0.9 |
13.0 |
GO:0000188 |
inactivation of MAPK activity(GO:0000188) |
| 0.9 |
3.6 |
GO:1903031 |
regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.9 |
2.6 |
GO:0021699 |
cerebellar Purkinje cell layer maturation(GO:0021691) cerebellar cortex maturation(GO:0021699) |
| 0.9 |
6.1 |
GO:0045200 |
establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.9 |
12.8 |
GO:0031507 |
heterochromatin assembly(GO:0031507) |
| 0.8 |
15.3 |
GO:0001574 |
ganglioside biosynthetic process(GO:0001574) |
| 0.8 |
3.3 |
GO:0060017 |
parathyroid gland development(GO:0060017) |
| 0.8 |
0.8 |
GO:0061324 |
cardiac right atrium morphogenesis(GO:0003213) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901963) |
| 0.8 |
2.4 |
GO:0000430 |
regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.8 |
8.1 |
GO:0015697 |
quaternary ammonium group transport(GO:0015697) |
| 0.8 |
4.8 |
GO:1900028 |
negative regulation of ruffle assembly(GO:1900028) |
| 0.8 |
3.2 |
GO:1901475 |
pyruvate transmembrane transport(GO:1901475) |
| 0.8 |
12.0 |
GO:2000114 |
regulation of establishment of cell polarity(GO:2000114) |
| 0.8 |
3.2 |
GO:0060709 |
glycogen cell differentiation involved in embryonic placenta development(GO:0060709) |
| 0.8 |
17.6 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.8 |
12.8 |
GO:0050832 |
defense response to fungus(GO:0050832) |
| 0.8 |
2.4 |
GO:0090285 |
regulation of protein glycosylation in Golgi(GO:0090283) negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.8 |
1.6 |
GO:0036216 |
response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.8 |
2.4 |
GO:0006592 |
ornithine biosynthetic process(GO:0006592) |
| 0.8 |
2.3 |
GO:0045404 |
positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.7 |
3.7 |
GO:0046880 |
progesterone secretion(GO:0042701) regulation of follicle-stimulating hormone secretion(GO:0046880) follicle-stimulating hormone secretion(GO:0046884) |
| 0.7 |
7.2 |
GO:0098903 |
regulation of membrane repolarization during action potential(GO:0098903) |
| 0.7 |
11.5 |
GO:0007216 |
G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.7 |
1.4 |
GO:0060025 |
regulation of synaptic activity(GO:0060025) |
| 0.7 |
5.7 |
GO:0046543 |
development of secondary female sexual characteristics(GO:0046543) |
| 0.7 |
11.9 |
GO:0021535 |
cell migration in hindbrain(GO:0021535) |
| 0.7 |
2.1 |
GO:0045054 |
constitutive secretory pathway(GO:0045054) |
| 0.7 |
28.1 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.7 |
2.7 |
GO:0006867 |
asparagine transport(GO:0006867) glutamine transport(GO:0006868) |
| 0.7 |
6.8 |
GO:0090557 |
establishment of endothelial intestinal barrier(GO:0090557) |
| 0.7 |
4.0 |
GO:0006177 |
GMP biosynthetic process(GO:0006177) |
| 0.7 |
7.9 |
GO:0038180 |
nerve growth factor signaling pathway(GO:0038180) |
| 0.7 |
3.3 |
GO:0036438 |
maintenance of lens transparency(GO:0036438) |
| 0.6 |
12.7 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.6 |
2.5 |
GO:1902268 |
negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.6 |
2.5 |
GO:0001920 |
negative regulation of receptor recycling(GO:0001920) |
| 0.6 |
3.1 |
GO:0070164 |
negative regulation of adiponectin secretion(GO:0070164) |
| 0.6 |
8.0 |
GO:0051481 |
negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.6 |
3.1 |
GO:1903361 |
protein localization to basolateral plasma membrane(GO:1903361) |
| 0.6 |
5.5 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.6 |
4.2 |
GO:0072176 |
nephric duct development(GO:0072176) nephric duct morphogenesis(GO:0072178) |
| 0.6 |
3.0 |
GO:0090286 |
cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.6 |
16.6 |
GO:0016242 |
negative regulation of macroautophagy(GO:0016242) |
| 0.6 |
8.9 |
GO:2001275 |
positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.6 |
1.7 |
GO:0035973 |
aggrephagy(GO:0035973) |
| 0.6 |
0.6 |
GO:0097709 |
connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.6 |
19.0 |
GO:0003301 |
physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.6 |
2.3 |
GO:0009212 |
dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.6 |
7.9 |
GO:0045161 |
neuronal ion channel clustering(GO:0045161) |
| 0.6 |
5.0 |
GO:0010763 |
positive regulation of fibroblast migration(GO:0010763) |
| 0.5 |
5.5 |
GO:1900112 |
regulation of histone H3-K9 trimethylation(GO:1900112) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.5 |
1.6 |
GO:0051823 |
regulation of synapse structural plasticity(GO:0051823) |
| 0.5 |
3.8 |
GO:0090091 |
microspike assembly(GO:0030035) regulation of microvillus assembly(GO:0032534) positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.5 |
2.1 |
GO:1904694 |
negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.5 |
36.9 |
GO:0000045 |
autophagosome assembly(GO:0000045) |
| 0.5 |
2.6 |
GO:0046501 |
protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.5 |
3.5 |
GO:1904217 |
regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.5 |
2.5 |
GO:0050912 |
detection of chemical stimulus involved in sensory perception(GO:0050907) detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.5 |
2.0 |
GO:1903265 |
positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.5 |
2.9 |
GO:1903056 |
mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.5 |
3.4 |
GO:0035887 |
aortic smooth muscle cell differentiation(GO:0035887) |
| 0.5 |
2.4 |
GO:1902775 |
mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.5 |
2.3 |
GO:0051012 |
microtubule sliding(GO:0051012) negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.5 |
0.9 |
GO:0045343 |
MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 0.5 |
3.2 |
GO:1900194 |
negative regulation of oocyte maturation(GO:1900194) |
| 0.5 |
3.2 |
GO:0030828 |
positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.5 |
12.9 |
GO:0060669 |
embryonic placenta morphogenesis(GO:0060669) |
| 0.5 |
0.5 |
GO:2000646 |
positive regulation of receptor catabolic process(GO:2000646) |
| 0.5 |
2.3 |
GO:0051013 |
microtubule severing(GO:0051013) |
| 0.4 |
10.3 |
GO:0021799 |
cerebral cortex radially oriented cell migration(GO:0021799) |
| 0.4 |
4.9 |
GO:0051764 |
actin crosslink formation(GO:0051764) |
| 0.4 |
3.6 |
GO:0042769 |
DNA damage response, detection of DNA damage(GO:0042769) |
| 0.4 |
2.2 |
GO:0006527 |
arginine catabolic process(GO:0006527) |
| 0.4 |
0.9 |
GO:2000271 |
positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.4 |
1.3 |
GO:0021698 |
cerebellar cortex structural organization(GO:0021698) |
| 0.4 |
2.6 |
GO:0033148 |
positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.4 |
7.7 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 0.4 |
5.1 |
GO:0060080 |
inhibitory postsynaptic potential(GO:0060080) |
| 0.4 |
9.7 |
GO:0002091 |
negative regulation of receptor internalization(GO:0002091) |
| 0.4 |
10.5 |
GO:0095500 |
acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.4 |
1.7 |
GO:0007023 |
post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.4 |
2.1 |
GO:0046208 |
spermine catabolic process(GO:0046208) |
| 0.4 |
4.1 |
GO:2000310 |
regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.4 |
11.1 |
GO:0007212 |
dopamine receptor signaling pathway(GO:0007212) |
| 0.4 |
11.4 |
GO:0009299 |
mRNA transcription(GO:0009299) |
| 0.4 |
2.0 |
GO:0070934 |
CRD-mediated mRNA stabilization(GO:0070934) |
| 0.4 |
1.2 |
GO:1900149 |
positive regulation of Schwann cell migration(GO:1900149) |
| 0.4 |
2.4 |
GO:0016338 |
calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.4 |
24.0 |
GO:0021954 |
central nervous system neuron development(GO:0021954) |
| 0.4 |
5.2 |
GO:0010758 |
regulation of macrophage chemotaxis(GO:0010758) |
| 0.4 |
0.4 |
GO:0007172 |
signal complex assembly(GO:0007172) negative regulation of synapse assembly(GO:0051964) |
| 0.4 |
1.6 |
GO:0034145 |
positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.4 |
0.8 |
GO:0000965 |
mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.4 |
6.3 |
GO:0099601 |
regulation of neurotransmitter receptor activity(GO:0099601) regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.4 |
2.7 |
GO:1901339 |
regulation of store-operated calcium channel activity(GO:1901339) |
| 0.4 |
6.9 |
GO:0071625 |
vocalization behavior(GO:0071625) |
| 0.4 |
1.9 |
GO:0009052 |
pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.4 |
6.1 |
GO:1902187 |
negative regulation of viral release from host cell(GO:1902187) |
| 0.4 |
3.1 |
GO:0045760 |
positive regulation of action potential(GO:0045760) |
| 0.4 |
8.0 |
GO:0060384 |
innervation(GO:0060384) |
| 0.4 |
1.9 |
GO:0021993 |
initiation of neural tube closure(GO:0021993) |
| 0.4 |
1.1 |
GO:1904417 |
regulation of xenophagy(GO:1904415) positive regulation of xenophagy(GO:1904417) |
| 0.4 |
1.1 |
GO:0045109 |
intermediate filament organization(GO:0045109) |
| 0.4 |
2.6 |
GO:0090073 |
positive regulation of protein homodimerization activity(GO:0090073) |
| 0.4 |
3.7 |
GO:0045671 |
negative regulation of osteoclast differentiation(GO:0045671) |
| 0.4 |
0.7 |
GO:0031133 |
regulation of axon diameter(GO:0031133) |
| 0.4 |
10.4 |
GO:0090200 |
positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.4 |
1.8 |
GO:0003383 |
apical constriction(GO:0003383) |
| 0.4 |
1.8 |
GO:0090666 |
scaRNA localization to Cajal body(GO:0090666) |
| 0.4 |
8.0 |
GO:0008053 |
mitochondrial fusion(GO:0008053) |
| 0.4 |
9.4 |
GO:0007617 |
mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.4 |
1.8 |
GO:0038032 |
termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.4 |
20.3 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.4 |
11.0 |
GO:0050919 |
negative chemotaxis(GO:0050919) |
| 0.4 |
3.5 |
GO:0022027 |
interkinetic nuclear migration(GO:0022027) |
| 0.3 |
2.1 |
GO:0016198 |
axon choice point recognition(GO:0016198) |
| 0.3 |
3.1 |
GO:0035280 |
miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.3 |
1.0 |
GO:0090241 |
negative regulation of histone H4 acetylation(GO:0090241) |
| 0.3 |
2.0 |
GO:1903755 |
regulation of SUMO transferase activity(GO:1903182) positive regulation of SUMO transferase activity(GO:1903755) |
| 0.3 |
2.7 |
GO:0033623 |
regulation of integrin activation(GO:0033623) |
| 0.3 |
3.3 |
GO:0035235 |
ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.3 |
1.3 |
GO:0030220 |
platelet formation(GO:0030220) |
| 0.3 |
1.3 |
GO:0010726 |
positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.3 |
2.9 |
GO:0032927 |
positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.3 |
1.3 |
GO:0043651 |
linoleic acid metabolic process(GO:0043651) |
| 0.3 |
5.5 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.3 |
3.5 |
GO:0002098 |
tRNA wobble uridine modification(GO:0002098) |
| 0.3 |
3.2 |
GO:0034643 |
establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.3 |
1.0 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.3 |
4.8 |
GO:0046855 |
phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.3 |
1.3 |
GO:1903071 |
positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.3 |
2.8 |
GO:1900119 |
positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.3 |
5.1 |
GO:0021591 |
ventricular system development(GO:0021591) |
| 0.3 |
5.3 |
GO:0031115 |
negative regulation of microtubule polymerization(GO:0031115) |
| 0.3 |
1.8 |
GO:0060346 |
bone trabecula formation(GO:0060346) |
| 0.3 |
2.1 |
GO:0048742 |
regulation of skeletal muscle fiber development(GO:0048742) |
| 0.3 |
1.5 |
GO:1904690 |
regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.3 |
0.9 |
GO:0010992 |
ubiquitin homeostasis(GO:0010992) |
| 0.3 |
2.4 |
GO:0006265 |
DNA topological change(GO:0006265) embryonic cleavage(GO:0040016) |
| 0.3 |
3.8 |
GO:0007340 |
acrosome reaction(GO:0007340) |
| 0.3 |
17.3 |
GO:0002028 |
regulation of sodium ion transport(GO:0002028) |
| 0.3 |
0.9 |
GO:0035928 |
rRNA import into mitochondrion(GO:0035928) |
| 0.3 |
2.0 |
GO:0071243 |
cellular response to arsenic-containing substance(GO:0071243) |
| 0.3 |
2.3 |
GO:0045075 |
interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.3 |
1.7 |
GO:0006182 |
cGMP biosynthetic process(GO:0006182) |
| 0.3 |
9.3 |
GO:0007218 |
neuropeptide signaling pathway(GO:0007218) |
| 0.3 |
20.9 |
GO:0048814 |
regulation of dendrite morphogenesis(GO:0048814) |
| 0.3 |
8.2 |
GO:0046627 |
negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.3 |
1.1 |
GO:0048148 |
behavioral response to cocaine(GO:0048148) |
| 0.3 |
2.0 |
GO:0033184 |
positive regulation of histone ubiquitination(GO:0033184) |
| 0.3 |
1.9 |
GO:0033008 |
positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.3 |
1.7 |
GO:0006968 |
cellular defense response(GO:0006968) |
| 0.3 |
2.4 |
GO:0032688 |
negative regulation of interferon-beta production(GO:0032688) |
| 0.3 |
7.8 |
GO:0016578 |
histone deubiquitination(GO:0016578) |
| 0.3 |
0.5 |
GO:1904059 |
regulation of locomotor rhythm(GO:1904059) |
| 0.3 |
0.8 |
GO:0003348 |
cardiac endothelial cell differentiation(GO:0003348) epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.3 |
1.1 |
GO:0001831 |
trophectodermal cellular morphogenesis(GO:0001831) |
| 0.3 |
2.8 |
GO:1901621 |
negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.3 |
3.1 |
GO:1900026 |
positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.3 |
9.4 |
GO:0007520 |
myoblast fusion(GO:0007520) |
| 0.3 |
2.5 |
GO:0035024 |
negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.3 |
10.3 |
GO:0070527 |
platelet aggregation(GO:0070527) |
| 0.3 |
1.8 |
GO:0050908 |
detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.3 |
1.0 |
GO:0045876 |
positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.2 |
1.5 |
GO:0043983 |
histone H4-K12 acetylation(GO:0043983) |
| 0.2 |
0.7 |
GO:0001869 |
regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.2 |
1.7 |
GO:1901223 |
negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.2 |
2.1 |
GO:0046831 |
regulation of RNA export from nucleus(GO:0046831) |
| 0.2 |
1.7 |
GO:0010216 |
maintenance of DNA methylation(GO:0010216) |
| 0.2 |
0.2 |
GO:0071864 |
positive regulation of cell proliferation in bone marrow(GO:0071864) |
| 0.2 |
3.5 |
GO:0010507 |
negative regulation of autophagy(GO:0010507) |
| 0.2 |
3.5 |
GO:0032331 |
negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.2 |
2.8 |
GO:0031987 |
locomotion involved in locomotory behavior(GO:0031987) |
| 0.2 |
19.3 |
GO:0010506 |
regulation of autophagy(GO:0010506) |
| 0.2 |
8.9 |
GO:0046488 |
phosphatidylinositol metabolic process(GO:0046488) |
| 0.2 |
0.7 |
GO:0002553 |
histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.2 |
0.5 |
GO:0030240 |
skeletal muscle thin filament assembly(GO:0030240) |
| 0.2 |
4.9 |
GO:0043153 |
entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.2 |
7.3 |
GO:0015844 |
monoamine transport(GO:0015844) |
| 0.2 |
5.0 |
GO:0048512 |
circadian behavior(GO:0048512) |
| 0.2 |
10.9 |
GO:0070059 |
intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.2 |
12.4 |
GO:0010923 |
negative regulation of phosphatase activity(GO:0010923) |
| 0.2 |
0.4 |
GO:0072553 |
terminal button organization(GO:0072553) |
| 0.2 |
0.6 |
GO:2000616 |
negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.2 |
0.8 |
GO:0016259 |
selenocysteine metabolic process(GO:0016259) |
| 0.2 |
1.4 |
GO:0036265 |
RNA (guanine-N7)-methylation(GO:0036265) |
| 0.2 |
1.6 |
GO:0060837 |
blood vessel endothelial cell differentiation(GO:0060837) |
| 0.2 |
0.8 |
GO:0060040 |
retinal bipolar neuron differentiation(GO:0060040) |
| 0.2 |
0.8 |
GO:1904587 |
glycoprotein ERAD pathway(GO:0097466) response to glycoprotein(GO:1904587) |
| 0.2 |
1.0 |
GO:1903887 |
motile primary cilium assembly(GO:1903887) |
| 0.2 |
1.4 |
GO:0070102 |
interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.2 |
2.9 |
GO:0043045 |
DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.2 |
0.6 |
GO:0034310 |
primary alcohol catabolic process(GO:0034310) |
| 0.2 |
0.4 |
GO:0060166 |
olfactory pit development(GO:0060166) |
| 0.2 |
0.4 |
GO:0034729 |
histone H3-K79 methylation(GO:0034729) |
| 0.2 |
0.9 |
GO:0035405 |
histone-threonine phosphorylation(GO:0035405) |
| 0.2 |
1.3 |
GO:0051014 |
actin filament severing(GO:0051014) |
| 0.2 |
1.1 |
GO:0097368 |
establishment of Sertoli cell barrier(GO:0097368) |
| 0.2 |
7.1 |
GO:0048813 |
dendrite morphogenesis(GO:0048813) |
| 0.2 |
2.2 |
GO:0006614 |
SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.2 |
32.6 |
GO:0007018 |
microtubule-based movement(GO:0007018) |
| 0.2 |
0.8 |
GO:2001197 |
positive regulation of extracellular matrix assembly(GO:1901203) positive regulation of extracellular matrix organization(GO:1903055) regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.2 |
1.0 |
GO:2000794 |
regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000794) |
| 0.2 |
1.9 |
GO:0070914 |
UV-damage excision repair(GO:0070914) |
| 0.2 |
5.0 |
GO:0006890 |
retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.2 |
9.5 |
GO:0006487 |
protein N-linked glycosylation(GO:0006487) |
| 0.2 |
4.5 |
GO:0046329 |
negative regulation of JNK cascade(GO:0046329) |
| 0.2 |
5.5 |
GO:0097202 |
activation of cysteine-type endopeptidase activity(GO:0097202) |
| 0.2 |
1.5 |
GO:0007193 |
adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.1 |
2.1 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 |
3.2 |
GO:0043392 |
negative regulation of DNA binding(GO:0043392) |
| 0.1 |
1.0 |
GO:0018344 |
protein geranylgeranylation(GO:0018344) |
| 0.1 |
2.0 |
GO:0042462 |
eye photoreceptor cell development(GO:0042462) |
| 0.1 |
1.4 |
GO:0034551 |
respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 |
3.3 |
GO:0031648 |
protein destabilization(GO:0031648) |
| 0.1 |
0.8 |
GO:0051152 |
positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 |
0.6 |
GO:1901642 |
purine nucleoside transmembrane transport(GO:0015860) purine-containing compound transmembrane transport(GO:0072530) nucleoside transmembrane transport(GO:1901642) |
| 0.1 |
2.1 |
GO:0045947 |
negative regulation of translational initiation(GO:0045947) |
| 0.1 |
0.8 |
GO:0051694 |
pointed-end actin filament capping(GO:0051694) |
| 0.1 |
9.2 |
GO:0006612 |
protein targeting to membrane(GO:0006612) |
| 0.1 |
4.7 |
GO:0034605 |
cellular response to heat(GO:0034605) |
| 0.1 |
1.3 |
GO:0031468 |
nuclear envelope reassembly(GO:0031468) |
| 0.1 |
5.0 |
GO:0051591 |
response to cAMP(GO:0051591) |
| 0.1 |
0.4 |
GO:2000020 |
positive regulation of male gonad development(GO:2000020) |
| 0.1 |
1.4 |
GO:0050995 |
negative regulation of lipid catabolic process(GO:0050995) |
| 0.1 |
0.9 |
GO:0043982 |
histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 |
0.9 |
GO:0035092 |
sperm chromatin condensation(GO:0035092) |
| 0.1 |
4.5 |
GO:0043044 |
ATP-dependent chromatin remodeling(GO:0043044) |
| 0.1 |
0.5 |
GO:0010288 |
response to lead ion(GO:0010288) |
| 0.1 |
1.1 |
GO:0048305 |
immunoglobulin secretion(GO:0048305) |
| 0.1 |
1.8 |
GO:0006308 |
DNA catabolic process(GO:0006308) DNA-templated transcription, termination(GO:0006353) |
| 0.1 |
17.7 |
GO:0008654 |
phospholipid biosynthetic process(GO:0008654) |
| 0.1 |
0.4 |
GO:0048211 |
Golgi vesicle docking(GO:0048211) |
| 0.1 |
0.5 |
GO:0045663 |
positive regulation of myoblast differentiation(GO:0045663) |
| 0.1 |
0.6 |
GO:0070828 |
heterochromatin organization(GO:0070828) |
| 0.1 |
1.2 |
GO:0002227 |
innate immune response in mucosa(GO:0002227) |
| 0.1 |
1.6 |
GO:0006904 |
vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 |
0.7 |
GO:0098787 |
mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 |
4.0 |
GO:0045921 |
positive regulation of exocytosis(GO:0045921) |
| 0.1 |
0.9 |
GO:0035269 |
protein O-linked mannosylation(GO:0035269) |
| 0.1 |
0.2 |
GO:0032878 |
regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.1 |
0.8 |
GO:0021796 |
cerebral cortex regionalization(GO:0021796) |
| 0.1 |
1.0 |
GO:0070389 |
chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 |
0.4 |
GO:1903587 |
regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.1 |
10.9 |
GO:0051017 |
actin filament bundle assembly(GO:0051017) actin filament bundle organization(GO:0061572) |
| 0.1 |
1.5 |
GO:0007157 |
heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 |
0.2 |
GO:0019805 |
quinolinate biosynthetic process(GO:0019805) |
| 0.1 |
3.9 |
GO:0043154 |
negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.1 |
0.4 |
GO:0035063 |
nuclear speck organization(GO:0035063) |
| 0.1 |
1.5 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.1 |
0.9 |
GO:0002643 |
regulation of tolerance induction(GO:0002643) |
| 0.1 |
0.7 |
GO:0035767 |
endothelial cell chemotaxis(GO:0035767) |
| 0.1 |
1.5 |
GO:0042417 |
dopamine metabolic process(GO:0042417) |
| 0.1 |
2.7 |
GO:0000470 |
maturation of LSU-rRNA(GO:0000470) |
| 0.1 |
4.2 |
GO:0030433 |
ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.1 |
0.6 |
GO:1903608 |
protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 |
7.1 |
GO:0006334 |
nucleosome assembly(GO:0006334) |
| 0.1 |
0.3 |
GO:0048239 |
negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.1 |
2.1 |
GO:0006301 |
postreplication repair(GO:0006301) |
| 0.1 |
3.0 |
GO:0009410 |
response to xenobiotic stimulus(GO:0009410) |
| 0.1 |
4.1 |
GO:0006749 |
glutathione metabolic process(GO:0006749) |
| 0.1 |
1.4 |
GO:0048026 |
positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 |
0.4 |
GO:1900095 |
regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.1 |
1.3 |
GO:0060479 |
lung cell differentiation(GO:0060479) lung epithelial cell differentiation(GO:0060487) |
| 0.1 |
0.5 |
GO:2000210 |
positive regulation of anoikis(GO:2000210) |
| 0.1 |
2.1 |
GO:0008542 |
visual learning(GO:0008542) |
| 0.1 |
1.6 |
GO:0000027 |
ribosomal large subunit assembly(GO:0000027) |
| 0.1 |
1.3 |
GO:0048009 |
insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.1 |
6.5 |
GO:0000086 |
G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.1 |
2.3 |
GO:0018208 |
peptidyl-proline modification(GO:0018208) |
| 0.1 |
0.4 |
GO:2000251 |
positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 |
0.7 |
GO:0090190 |
positive regulation of mesonephros development(GO:0061213) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.1 |
4.4 |
GO:0030518 |
intracellular steroid hormone receptor signaling pathway(GO:0030518) |
| 0.1 |
0.9 |
GO:0001967 |
suckling behavior(GO:0001967) |
| 0.1 |
1.5 |
GO:0048013 |
ephrin receptor signaling pathway(GO:0048013) |
| 0.1 |
1.3 |
GO:0016180 |
snRNA processing(GO:0016180) |
| 0.1 |
0.5 |
GO:0051491 |
positive regulation of filopodium assembly(GO:0051491) |
| 0.1 |
0.3 |
GO:0048199 |
vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.1 |
1.0 |
GO:0071108 |
protein K48-linked deubiquitination(GO:0071108) |
| 0.1 |
3.5 |
GO:0035383 |
acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.1 |
1.0 |
GO:0006607 |
NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 |
0.4 |
GO:0043488 |
regulation of mRNA stability(GO:0043488) |
| 0.1 |
1.3 |
GO:0030317 |
sperm motility(GO:0030317) |
| 0.1 |
1.0 |
GO:0007608 |
sensory perception of smell(GO:0007608) |
| 0.1 |
0.3 |
GO:0001672 |
regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 |
0.5 |
GO:0032292 |
myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 |
0.5 |
GO:0035025 |
positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 |
0.1 |
GO:0045654 |
positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 |
2.7 |
GO:0048167 |
regulation of synaptic plasticity(GO:0048167) |
| 0.0 |
0.5 |
GO:0060218 |
hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 |
0.1 |
GO:0000707 |
meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 |
1.0 |
GO:0034314 |
Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 |
0.5 |
GO:0090161 |
Golgi ribbon formation(GO:0090161) |
| 0.0 |
0.4 |
GO:0010172 |
embryonic body morphogenesis(GO:0010172) |
| 0.0 |
0.3 |
GO:0006044 |
N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 |
0.1 |
GO:0014842 |
regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.0 |
0.5 |
GO:0030150 |
protein import into mitochondrial matrix(GO:0030150) |
| 0.0 |
0.8 |
GO:0003333 |
amino acid transmembrane transport(GO:0003333) |
| 0.0 |
1.3 |
GO:0051865 |
protein autoubiquitination(GO:0051865) |
| 0.0 |
0.5 |
GO:0042273 |
ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 |
0.1 |
GO:0006102 |
isocitrate metabolic process(GO:0006102) |
| 0.0 |
3.5 |
GO:0018105 |
peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 |
0.7 |
GO:1902850 |
mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.0 |
0.1 |
GO:0006420 |
arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 |
0.3 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.0 |
0.2 |
GO:0016601 |
Rac protein signal transduction(GO:0016601) |
| 0.0 |
0.1 |
GO:0016598 |
protein arginylation(GO:0016598) |
| 0.0 |
0.2 |
GO:0080111 |
DNA demethylation(GO:0080111) |
| 0.0 |
0.2 |
GO:0001825 |
blastocyst formation(GO:0001825) |
| 0.0 |
0.0 |
GO:0070236 |
negative regulation of activation-induced cell death of T cells(GO:0070236) |