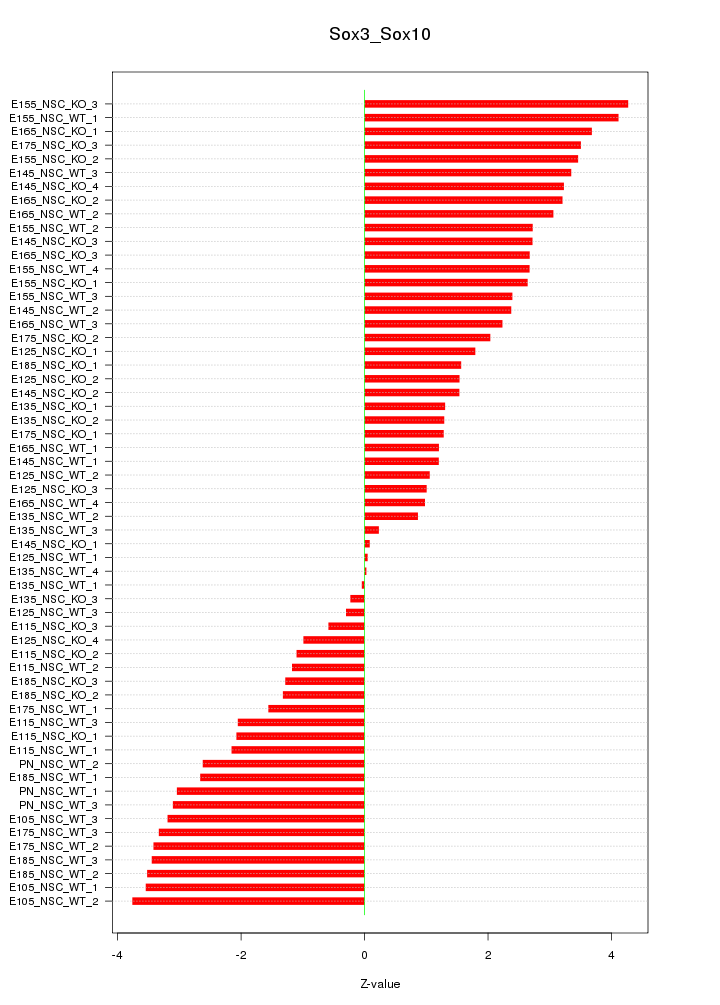
Motif ID: Sox3_Sox10
Z-value: 2.376
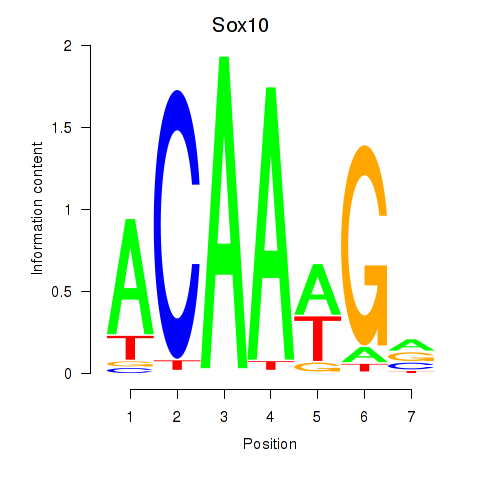
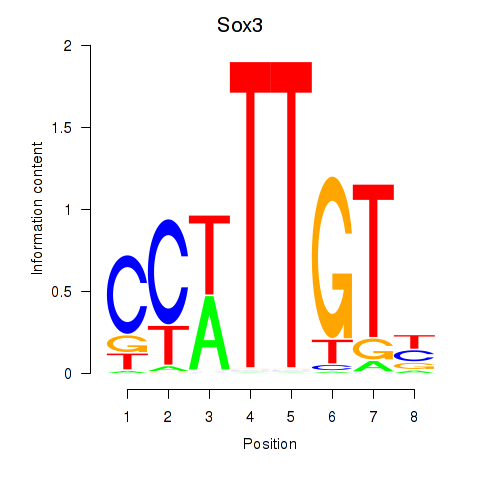
Transcription factors associated with Sox3_Sox10:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Sox10 | ENSMUSG00000033006.9 | Sox10 |
| Sox3 | ENSMUSG00000045179.8 | Sox3 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox10 | mm10_v2_chr15_-_79164477_79164496 | -0.48 | 1.4e-04 | Click! |
| Sox3 | mm10_v2_chrX_-_60893430_60893440 | -0.32 | 1.4e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.7 | 41.1 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 8.6 | 25.7 | GO:0042560 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 7.4 | 29.8 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 6.7 | 6.7 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 5.8 | 23.3 | GO:1904529 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 5.7 | 17.0 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 5.6 | 22.4 | GO:0032289 | central nervous system myelin formation(GO:0032289) cardiac cell fate specification(GO:0060912) |
| 5.4 | 16.1 | GO:0030827 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 4.9 | 24.4 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 4.6 | 106.0 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 4.4 | 30.5 | GO:0034047 | regulation of protein phosphatase type 2A activity(GO:0034047) |
| 4.3 | 69.4 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 4.1 | 49.0 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 4.1 | 12.2 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 4.1 | 16.2 | GO:0021764 | amygdala development(GO:0021764) |
| 4.0 | 4.0 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 3.9 | 11.8 | GO:0014877 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 3.9 | 19.6 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 3.9 | 11.7 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 3.6 | 14.3 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 3.5 | 14.1 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 3.2 | 9.6 | GO:0038095 | positive regulation of mast cell cytokine production(GO:0032765) Fc-epsilon receptor signaling pathway(GO:0038095) |
| 3.1 | 12.5 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 3.1 | 12.5 | GO:2001016 | positive regulation of skeletal muscle cell differentiation(GO:2001016) |
| 3.0 | 12.0 | GO:0003357 | noradrenergic neuron differentiation(GO:0003357) |
| 3.0 | 14.8 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 2.8 | 17.1 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 2.7 | 10.9 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 2.6 | 10.3 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 2.5 | 7.5 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 2.5 | 7.5 | GO:0021649 | vestibulocochlear nerve morphogenesis(GO:0021648) vestibulocochlear nerve structural organization(GO:0021649) ganglion morphogenesis(GO:0061552) dorsal root ganglion morphogenesis(GO:1904835) dorsal root ganglion development(GO:1990791) |
| 2.5 | 14.9 | GO:0060371 | regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) |
| 2.5 | 9.8 | GO:0003105 | negative regulation of glomerular filtration(GO:0003105) |
| 2.3 | 7.0 | GO:0070428 | regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 2.3 | 32.7 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 2.3 | 9.1 | GO:1990743 | protein sialylation(GO:1990743) |
| 2.2 | 17.5 | GO:0021546 | rhombomere development(GO:0021546) |
| 2.1 | 6.4 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 2.1 | 4.2 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 2.1 | 8.4 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 2.1 | 16.6 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 2.1 | 22.8 | GO:0086018 | SA node cell action potential(GO:0086015) SA node cell to atrial cardiac muscle cell signalling(GO:0086018) SA node cell to atrial cardiac muscle cell communication(GO:0086070) |
| 1.9 | 17.0 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 1.9 | 13.1 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 1.9 | 5.6 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 1.8 | 32.4 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) negative regulation of cyclase activity(GO:0031280) |
| 1.7 | 7.0 | GO:0032512 | regulation of protein phosphatase type 2B activity(GO:0032512) positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 1.7 | 1.7 | GO:0050883 | musculoskeletal movement, spinal reflex action(GO:0050883) |
| 1.7 | 5.2 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 1.7 | 8.6 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 1.7 | 12.0 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 1.7 | 5.0 | GO:0032650 | regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) interleukin-1 alpha secretion(GO:0050703) |
| 1.6 | 19.7 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 1.6 | 4.9 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 1.6 | 4.9 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 1.6 | 13.0 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 1.6 | 8.0 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 1.5 | 4.5 | GO:0060489 | epicardial cell to mesenchymal cell transition(GO:0003347) establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 1.5 | 2.9 | GO:0021933 | radial glia guided migration of cerebellar granule cell(GO:0021933) |
| 1.5 | 8.7 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 1.4 | 4.1 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 1.4 | 19.2 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 1.3 | 8.9 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 1.2 | 11.2 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 1.2 | 3.7 | GO:0007525 | somatic muscle development(GO:0007525) |
| 1.2 | 9.8 | GO:0019227 | neuronal action potential propagation(GO:0019227) detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) action potential propagation(GO:0098870) |
| 1.2 | 1.2 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 1.2 | 23.9 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 1.2 | 5.9 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 1.2 | 3.5 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 1.2 | 15.1 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 1.2 | 8.1 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 1.1 | 12.5 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 1.1 | 3.3 | GO:0017085 | response to insecticide(GO:0017085) |
| 1.1 | 8.8 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 1.1 | 17.5 | GO:1901629 | regulation of presynaptic membrane organization(GO:1901629) |
| 1.1 | 10.9 | GO:1900112 | regulation of histone H3-K9 trimethylation(GO:1900112) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 1.0 | 4.2 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 1.0 | 9.3 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 1.0 | 15.7 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 1.0 | 17.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 1.0 | 12.4 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.9 | 4.7 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.9 | 2.8 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.9 | 7.2 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.9 | 5.4 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.9 | 2.6 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.9 | 5.2 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.9 | 8.6 | GO:0071635 | negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.9 | 18.0 | GO:0048148 | behavioral response to cocaine(GO:0048148) |
| 0.8 | 4.2 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.8 | 3.3 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.8 | 7.3 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.8 | 6.4 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.8 | 2.4 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.8 | 3.1 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.8 | 3.9 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.8 | 2.3 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.7 | 8.1 | GO:0071225 | cellular response to muramyl dipeptide(GO:0071225) |
| 0.7 | 1.5 | GO:0003096 | renal sodium ion transport(GO:0003096) |
| 0.7 | 5.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.7 | 2.8 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.7 | 2.1 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.7 | 17.1 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.7 | 2.0 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.7 | 6.7 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.7 | 4.7 | GO:1905206 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) positive regulation of hydrogen peroxide-induced cell death(GO:1905206) |
| 0.7 | 7.2 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.7 | 9.8 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.6 | 1.3 | GO:0060025 | regulation of synaptic activity(GO:0060025) |
| 0.6 | 1.9 | GO:0021590 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.6 | 1.9 | GO:0071544 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.6 | 5.8 | GO:0034776 | response to histamine(GO:0034776) cellular response to histamine(GO:0071420) |
| 0.6 | 1.3 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.6 | 3.7 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.6 | 1.8 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.6 | 39.9 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.6 | 2.4 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.6 | 12.4 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.6 | 4.1 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.6 | 1.8 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.6 | 2.9 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.6 | 7.0 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.6 | 10.5 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.6 | 4.0 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.6 | 1.7 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.6 | 5.1 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.6 | 3.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.6 | 2.8 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.6 | 19.5 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.6 | 2.2 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.5 | 2.2 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.5 | 3.8 | GO:0014848 | urinary bladder smooth muscle contraction(GO:0014832) urinary tract smooth muscle contraction(GO:0014848) |
| 0.5 | 3.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.5 | 19.4 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.5 | 3.2 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.5 | 5.3 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.5 | 2.1 | GO:0034145 | positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.5 | 1.6 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.5 | 1.6 | GO:0002035 | brain renin-angiotensin system(GO:0002035) |
| 0.5 | 1.6 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.5 | 1.0 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.5 | 3.1 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.5 | 5.6 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.5 | 4.6 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.5 | 4.5 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.5 | 1.5 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.5 | 1.5 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.5 | 2.5 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.5 | 3.5 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.5 | 5.4 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.5 | 2.4 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.5 | 20.4 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.5 | 1.0 | GO:0046098 | regulation of primitive erythrocyte differentiation(GO:0010725) guanine metabolic process(GO:0046098) |
| 0.5 | 0.5 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.5 | 1.4 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.5 | 31.9 | GO:0021954 | central nervous system neuron development(GO:0021954) |
| 0.5 | 0.5 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.5 | 3.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.5 | 29.8 | GO:0000045 | autophagosome assembly(GO:0000045) |
| 0.5 | 3.7 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.4 | 16.2 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.4 | 12.5 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.4 | 1.8 | GO:0045213 | neurotransmitter receptor metabolic process(GO:0045213) |
| 0.4 | 4.4 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.4 | 7.0 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.4 | 2.6 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.4 | 12.7 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.4 | 4.8 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.4 | 11.3 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.4 | 12.2 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.4 | 0.8 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.4 | 1.6 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.4 | 0.8 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.4 | 1.2 | GO:0035984 | response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.4 | 2.8 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.4 | 1.9 | GO:0050912 | detection of chemical stimulus involved in sensory perception(GO:0050907) detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.4 | 54.1 | GO:0030041 | actin filament polymerization(GO:0030041) |
| 0.4 | 1.9 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.4 | 1.4 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.4 | 4.2 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.3 | 6.2 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.3 | 2.0 | GO:1902993 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.3 | 4.0 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.3 | 6.0 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.3 | 6.4 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.3 | 2.9 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.3 | 1.3 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.3 | 1.6 | GO:0000022 | mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.3 | 1.6 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.3 | 21.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.3 | 0.6 | GO:1903351 | response to dopamine(GO:1903350) cellular response to dopamine(GO:1903351) regulation of sodium:potassium-exchanging ATPase activity(GO:1903406) |
| 0.3 | 6.9 | GO:0095500 | acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.3 | 6.8 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.3 | 0.6 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.3 | 1.2 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.3 | 1.5 | GO:0051012 | microtubule sliding(GO:0051012) negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.3 | 2.4 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.3 | 6.9 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.3 | 0.9 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.3 | 1.5 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.3 | 0.9 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.3 | 1.8 | GO:2000096 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.3 | 5.5 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.3 | 1.1 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.3 | 1.1 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.3 | 2.8 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.3 | 7.3 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.3 | 0.3 | GO:2000646 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) positive regulation of receptor catabolic process(GO:2000646) |
| 0.3 | 1.3 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.3 | 31.1 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.3 | 1.3 | GO:0060903 | regulation of meiosis I(GO:0060631) positive regulation of meiosis I(GO:0060903) |
| 0.3 | 1.8 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.3 | 0.3 | GO:2000830 | vacuolar phosphate transport(GO:0007037) vitamin D3 metabolic process(GO:0070640) regulation of parathyroid hormone secretion(GO:2000828) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.3 | 1.3 | GO:0071476 | cellular hypotonic response(GO:0071476) |
| 0.2 | 16.5 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.2 | 0.7 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.2 | 2.7 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.2 | 5.3 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 1.2 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.2 | 0.9 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.2 | 0.7 | GO:0051683 | establishment of Golgi localization(GO:0051683) |
| 0.2 | 0.5 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.2 | 5.3 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.2 | 4.0 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.2 | 0.6 | GO:0009838 | abscission(GO:0009838) |
| 0.2 | 0.8 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.2 | 3.3 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.2 | 1.2 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.2 | 0.4 | GO:0051580 | regulation of neurotransmitter uptake(GO:0051580) |
| 0.2 | 18.3 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.2 | 3.6 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.2 | 0.8 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.2 | 2.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.2 | 26.5 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.2 | 34.4 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.2 | 1.5 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.2 | 12.2 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.2 | 2.8 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.2 | 3.5 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.2 | 1.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.2 | 2.1 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.2 | 0.9 | GO:0072429 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.2 | 1.2 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.2 | 0.7 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.2 | 8.6 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.2 | 2.2 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.2 | 6.0 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.2 | 1.2 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.2 | 14.1 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.2 | 5.1 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.2 | 0.5 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.2 | 0.3 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.2 | 1.6 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.2 | 4.4 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.2 | 0.6 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.2 | 1.5 | GO:1901409 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.2 | 2.7 | GO:0002437 | inflammatory response to antigenic stimulus(GO:0002437) |
| 0.2 | 3.8 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.2 | 0.5 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.2 | 3.3 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.2 | 1.9 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.2 | 1.2 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.2 | 6.7 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.2 | 0.3 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.1 | 0.7 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 0.3 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 | 4.3 | GO:0008542 | visual behavior(GO:0007632) visual learning(GO:0008542) |
| 0.1 | 2.3 | GO:0001783 | B cell apoptotic process(GO:0001783) |
| 0.1 | 1.3 | GO:0015838 | amino-acid betaine transport(GO:0015838) |
| 0.1 | 1.1 | GO:0042095 | interferon-gamma biosynthetic process(GO:0042095) |
| 0.1 | 1.0 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.1 | 0.6 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 1.9 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.1 | 6.7 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 0.2 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.1 | 0.5 | GO:0006867 | asparagine transport(GO:0006867) glutamine transport(GO:0006868) |
| 0.1 | 1.3 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 3.5 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 0.9 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.1 | 0.3 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.1 | 3.8 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 0.9 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 1.0 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.1 | 2.5 | GO:0060291 | long-term synaptic potentiation(GO:0060291) |
| 0.1 | 2.6 | GO:0015844 | monoamine transport(GO:0015844) |
| 0.1 | 0.4 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 1.7 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.1 | 1.6 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 0.3 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.8 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 1.6 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.1 | 1.2 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 2.1 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.1 | 1.0 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 0.5 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 3.5 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.1 | 0.8 | GO:0050995 | negative regulation of lipid catabolic process(GO:0050995) |
| 0.1 | 3.0 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.1 | 1.9 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.1 | 1.3 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.1 | 0.4 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.1 | 0.2 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.1 | 0.5 | GO:0046036 | GTP biosynthetic process(GO:0006183) CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.1 | 0.5 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.1 | 2.5 | GO:0048167 | regulation of synaptic plasticity(GO:0048167) |
| 0.1 | 0.4 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 0.7 | GO:0033198 | response to ATP(GO:0033198) |
| 0.1 | 0.3 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.5 | GO:0071624 | positive regulation of granulocyte chemotaxis(GO:0071624) positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.1 | 2.2 | GO:0050777 | negative regulation of immune response(GO:0050777) |
| 0.0 | 0.5 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 1.1 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.7 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.2 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.0 | 0.2 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.6 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 10.7 | GO:0007017 | microtubule-based process(GO:0007017) |
| 0.0 | 0.2 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.4 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 1.9 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 0.7 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.8 | GO:0006906 | vesicle fusion(GO:0006906) |
| 0.0 | 1.6 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.8 | GO:0048536 | spleen development(GO:0048536) |
| 0.0 | 4.5 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 1.4 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.8 | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.0 | 0.7 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.3 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 1.1 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.5 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:0045618 | lipoxygenase pathway(GO:0019372) positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.0 | 0.2 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.0 | 0.1 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.3 | GO:0048477 | oogenesis(GO:0048477) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.0 | 70.0 | GO:0045298 | tubulin complex(GO:0045298) |
| 6.0 | 24.1 | GO:0044307 | dendritic branch(GO:0044307) |
| 4.9 | 44.2 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 3.6 | 10.9 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 3.2 | 19.4 | GO:1990635 | proximal dendrite(GO:1990635) |
| 3.1 | 12.5 | GO:0043259 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) |
| 3.0 | 75.9 | GO:0071565 | nBAF complex(GO:0071565) |
| 2.5 | 39.3 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 2.3 | 18.3 | GO:0001520 | outer dense fiber(GO:0001520) |
| 2.2 | 6.6 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 1.9 | 17.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 1.8 | 34.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 1.8 | 16.1 | GO:0044327 | dendritic spine head(GO:0044327) |
| 1.8 | 14.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 1.7 | 6.7 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 1.7 | 6.7 | GO:1902737 | dendritic filopodium(GO:1902737) |
| 1.6 | 7.9 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 1.5 | 11.9 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 1.4 | 29.8 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 1.4 | 8.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 1.4 | 5.5 | GO:0090537 | CERF complex(GO:0090537) |
| 1.3 | 57.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 1.2 | 15.1 | GO:0016342 | catenin complex(GO:0016342) |
| 1.1 | 5.7 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 1.1 | 4.5 | GO:0060187 | cell pole(GO:0060187) |
| 1.0 | 12.4 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.9 | 45.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.9 | 4.6 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.9 | 9.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.9 | 6.0 | GO:0005638 | lamin filament(GO:0005638) |
| 0.8 | 10.0 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.8 | 4.9 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.8 | 3.2 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.8 | 2.3 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.7 | 16.8 | GO:0031430 | M band(GO:0031430) |
| 0.7 | 4.9 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.7 | 7.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.7 | 2.8 | GO:0031673 | H zone(GO:0031673) |
| 0.7 | 15.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.7 | 5.9 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.6 | 12.0 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.6 | 7.9 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.6 | 1.8 | GO:0044308 | axonal spine(GO:0044308) |
| 0.6 | 5.3 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.6 | 4.0 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.6 | 1.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.6 | 6.8 | GO:0042581 | specific granule(GO:0042581) |
| 0.5 | 26.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.5 | 35.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.5 | 21.4 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.5 | 50.1 | GO:0005884 | actin filament(GO:0005884) |
| 0.5 | 1.6 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.5 | 23.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.5 | 5.6 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.5 | 4.0 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.5 | 6.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.5 | 11.8 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.5 | 33.8 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.5 | 4.7 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.5 | 14.1 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.5 | 28.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.5 | 8.6 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.4 | 6.0 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.4 | 2.6 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.4 | 5.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.4 | 209.2 | GO:0030425 | dendrite(GO:0030425) |
| 0.4 | 6.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.4 | 1.4 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.4 | 3.5 | GO:0031988 | membrane-bounded vesicle(GO:0031988) |
| 0.3 | 17.6 | GO:0005776 | autophagosome(GO:0005776) |
| 0.3 | 19.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.3 | 6.6 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.3 | 4.4 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.3 | 2.8 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.3 | 1.4 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.3 | 12.7 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.3 | 2.6 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.3 | 1.5 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.3 | 3.3 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.2 | 1.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.2 | 6.8 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.2 | 8.6 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.2 | 4.3 | GO:0043679 | axon terminus(GO:0043679) |
| 0.2 | 2.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.2 | 0.8 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.2 | 3.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.2 | 2.6 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.2 | 0.9 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.2 | 4.7 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.2 | 3.1 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.2 | 0.7 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.2 | 0.9 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.2 | 0.9 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.2 | 6.0 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.2 | 10.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.2 | 1.6 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.2 | 19.9 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.2 | 2.5 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.2 | 4.5 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.2 | 1.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.2 | 2.2 | GO:0098797 | plasma membrane protein complex(GO:0098797) |
| 0.1 | 2.5 | GO:0031253 | cell projection membrane(GO:0031253) |
| 0.1 | 8.0 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 9.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 2.6 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 7.7 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.1 | 0.8 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 1.6 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.2 | GO:0051286 | cell tip(GO:0051286) |
| 0.1 | 0.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 6.9 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 0.8 | GO:0044298 | cell body membrane(GO:0044298) |
| 0.1 | 0.8 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 0.3 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.1 | 1.6 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.4 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 2.9 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 3.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 0.7 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 1.6 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 1.3 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 2.6 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.1 | 6.7 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 3.1 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 2.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 5.5 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 0.3 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 3.8 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 0.5 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.9 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.2 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 2.0 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.3 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 0.1 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 1.5 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 1.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 2.3 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.3 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 11.1 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.1 | GO:0044437 | vacuolar membrane(GO:0005774) vacuolar part(GO:0044437) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.4 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.5 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.3 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.3 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.6 | 25.7 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 6.3 | 50.5 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 5.6 | 28.2 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 5.2 | 20.6 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 3.4 | 17.0 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 3.2 | 72.9 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 3.0 | 12.2 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 3.0 | 71.4 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 2.9 | 17.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 2.4 | 16.9 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 2.4 | 12.0 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 2.2 | 9.0 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 2.2 | 30.5 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 2.2 | 8.6 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 2.1 | 6.4 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 2.1 | 14.9 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 2.0 | 26.5 | GO:0031005 | filamin binding(GO:0031005) |
| 2.0 | 14.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 2.0 | 31.5 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 1.9 | 26.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 1.8 | 5.5 | GO:0070615 | nucleosome-dependent ATPase activity(GO:0070615) |
| 1.8 | 12.7 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 1.8 | 14.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 1.8 | 17.8 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 1.7 | 8.4 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 1.6 | 8.0 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 1.6 | 4.8 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 1.6 | 7.9 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 1.6 | 26.9 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 1.6 | 12.5 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 1.6 | 17.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 1.5 | 4.6 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 1.5 | 31.1 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 1.5 | 8.7 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 1.4 | 11.2 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 1.4 | 4.1 | GO:0043912 | D-lysine oxidase activity(GO:0043912) |
| 1.3 | 4.0 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 1.3 | 7.9 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 1.3 | 3.9 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 1.2 | 3.5 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 1.2 | 9.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 1.2 | 3.5 | GO:0050827 | toxin receptor binding(GO:0050827) |
| 1.1 | 6.8 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 1.1 | 12.4 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 1.1 | 4.4 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 1.1 | 24.0 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 1.1 | 7.5 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 1.0 | 3.1 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
| 1.0 | 22.8 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 1.0 | 13.3 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 1.0 | 8.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 1.0 | 2.9 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 1.0 | 5.7 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.9 | 2.8 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.9 | 11.2 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.9 | 3.7 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.9 | 14.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.9 | 10.6 | GO:0022842 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.9 | 26.4 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.9 | 2.6 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.8 | 12.7 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.8 | 6.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.8 | 5.7 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.8 | 3.2 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.8 | 7.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.8 | 23.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.8 | 18.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.8 | 3.2 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.8 | 9.5 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.8 | 9.8 | GO:0005522 | profilin binding(GO:0005522) |
| 0.7 | 3.0 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.7 | 4.5 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.7 | 2.2 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.7 | 4.2 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.7 | 9.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.7 | 20.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.7 | 24.1 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.7 | 16.5 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.7 | 6.0 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.7 | 2.6 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.7 | 32.0 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.6 | 1.9 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.6 | 9.6 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.6 | 21.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.6 | 8.1 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.6 | 37.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.6 | 3.9 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.5 | 1.6 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.5 | 5.8 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.5 | 35.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.5 | 4.6 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.5 | 16.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.5 | 1.5 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.5 | 5.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.5 | 83.1 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.5 | 5.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.5 | 2.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.4 | 1.3 | GO:0046592 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.4 | 4.4 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.4 | 9.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.4 | 5.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.4 | 6.7 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.4 | 3.7 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.4 | 1.6 | GO:0071532 | ornithine decarboxylase inhibitor activity(GO:0008073) ankyrin repeat binding(GO:0071532) |
| 0.4 | 1.6 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.4 | 14.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.4 | 8.9 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.4 | 10.6 | GO:0004120 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) photoreceptor cyclic-nucleotide phosphodiesterase activity(GO:0004120) 7,8-dihydro-D-neopterin 2',3'-cyclic phosphate phosphodiesterase activity(GO:0044688) inositol phosphosphingolipid phospholipase activity(GO:0052712) inositol phosphorylceramide phospholipase activity(GO:0052713) mannosyl-inositol phosphorylceramide phospholipase activity(GO:0052714) mannosyl-diinositol phosphorylceramide phospholipase activity(GO:0052715) |
| 0.4 | 3.8 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.4 | 4.2 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.4 | 22.5 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.4 | 6.8 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.4 | 7.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.3 | 6.6 | GO:0043747 | protein-N-terminal asparagine amidohydrolase activity(GO:0008418) UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase activity(GO:0008759) iprodione amidohydrolase activity(GO:0018748) (3,5-dichlorophenylurea)acetate amidohydrolase activity(GO:0018749) 4'-(2-hydroxyisopropyl)phenylurea amidohydrolase activity(GO:0034571) didemethylisoproturon amidohydrolase activity(GO:0034573) N-isopropylacetanilide amidohydrolase activity(GO:0034576) N-cyclohexylformamide amidohydrolase activity(GO:0034781) isonicotinic acid hydrazide hydrolase activity(GO:0034876) cis-aconitamide amidase activity(GO:0034882) gamma-N-formylaminovinylacetate hydrolase activity(GO:0034885) N2-acetyl-L-lysine deacetylase activity(GO:0043747) O-succinylbenzoate synthase activity(GO:0043748) indoleacetamide hydrolase activity(GO:0043864) N-acetylcitrulline deacetylase activity(GO:0043909) N-acetylgalactosamine-6-phosphate deacetylase activity(GO:0047419) diacetylchitobiose deacetylase activity(GO:0052773) chitooligosaccharide deacetylase activity(GO:0052790) |
| 0.3 | 11.9 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.3 | 6.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.3 | 1.0 | GO:0015173 | hydrogen:amino acid symporter activity(GO:0005280) aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.3 | 1.6 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.3 | 2.9 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.3 | 1.9 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.3 | 1.3 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.3 | 10.8 | GO:0017022 | myosin binding(GO:0017022) |
| 0.3 | 13.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.3 | 1.9 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.3 | 4.6 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.3 | 1.2 | GO:2001070 | glycerophosphocholine phosphodiesterase activity(GO:0047389) starch binding(GO:2001070) |
| 0.3 | 1.5 | GO:0099528 | G-protein coupled neurotransmitter receptor activity(GO:0099528) |
| 0.3 | 1.2 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.3 | 2.6 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.3 | 86.3 | GO:0043774 | UDP-N-acetylmuramoylalanyl-D-glutamyl-2,6-diaminopimelate-D-alanyl-D-alanine ligase activity(GO:0008766) ribosomal S6-glutamic acid ligase activity(GO:0018169) coenzyme F420-0 gamma-glutamyl ligase activity(GO:0043773) coenzyme F420-2 alpha-glutamyl ligase activity(GO:0043774) protein-glycine ligase activity(GO:0070735) protein-glycine ligase activity, initiating(GO:0070736) protein-glycine ligase activity, elongating(GO:0070737) tubulin-glycine ligase activity(GO:0070738) |
| 0.3 | 4.9 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.3 | 1.7 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.3 | 3.6 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) lipoprotein particle receptor binding(GO:0070325) |
| 0.3 | 4.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.3 | 1.9 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.3 | 4.0 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.3 | 3.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.3 | 4.5 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.3 | 1.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.3 | 4.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.3 | 4.8 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.2 | 0.2 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.2 | 1.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.2 | 5.1 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.2 | 4.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 5.7 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 0.7 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.2 | 1.8 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 4.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.2 | 12.0 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.2 | 1.3 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.2 | 6.5 | GO:0018602 | sulfonate dioxygenase activity(GO:0000907) 2,4-dichlorophenoxyacetate alpha-ketoglutarate dioxygenase activity(GO:0018602) hypophosphite dioxygenase activity(GO:0034792) gibberellin 2-beta-dioxygenase activity(GO:0045543) C-19 gibberellin 2-beta-dioxygenase activity(GO:0052634) C-20 gibberellin 2-beta-dioxygenase activity(GO:0052635) |
| 0.2 | 5.0 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.2 | 5.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.2 | 1.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.2 | 1.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.2 | 3.8 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.2 | 0.8 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.2 | 4.8 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.2 | 8.2 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.2 | 0.6 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.2 | 0.7 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.2 | 23.6 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.2 | 3.0 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.2 | 55.3 | GO:0003779 | actin binding(GO:0003779) |
| 0.2 | 5.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.2 | 2.8 | GO:0042805 | actinin binding(GO:0042805) |
| 0.2 | 1.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.2 | 1.8 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.2 | 0.8 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.2 | 2.0 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.2 | 0.5 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.2 | 0.9 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 0.9 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 0.9 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 2.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 5.9 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.1 | 4.9 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 15.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 3.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 12.9 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.1 | 7.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 0.4 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 1.7 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.1 | 1.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.5 | GO:0015186 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 1.1 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.1 | 0.5 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.1 | 1.0 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 10.1 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 4.8 | GO:0061733 | peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.1 | 4.7 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 2.3 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 7.2 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 1.8 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 14.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 1.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 1.9 | GO:0034930 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) heparan sulfate 2-O-sulfotransferase activity(GO:0004394) HNK-1 sulfotransferase activity(GO:0016232) heparan sulfate 6-O-sulfotransferase activity(GO:0017095) trans-9R,10R-dihydrodiolphenanthrene sulfotransferase activity(GO:0018721) 1-phenanthrol sulfotransferase activity(GO:0018722) 3-phenanthrol sulfotransferase activity(GO:0018723) 4-phenanthrol sulfotransferase activity(GO:0018724) trans-3,4-dihydrodiolphenanthrene sulfotransferase activity(GO:0018725) 9-phenanthrol sulfotransferase activity(GO:0018726) 2-phenanthrol sulfotransferase activity(GO:0018727) phenanthrol sulfotransferase activity(GO:0019111) 1-hydroxypyrene sulfotransferase activity(GO:0034930) proteoglycan sulfotransferase activity(GO:0050698) cholesterol sulfotransferase activity(GO:0051922) hydroxyjasmonate sulfotransferase activity(GO:0080131) |
| 0.1 | 4.0 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 4.1 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 16.3 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 0.4 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.1 | 1.3 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 0.5 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 2.4 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 1.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 1.0 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 2.0 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 0.5 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 0.7 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 2.2 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 1.0 | GO:0005496 | steroid binding(GO:0005496) |
| 0.1 | 5.7 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.5 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 1.9 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.2 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.5 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.9 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 1.0 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.7 | GO:0032934 | cholesterol binding(GO:0015485) sterol binding(GO:0032934) |
| 0.0 | 0.1 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.5 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 2.6 | GO:0042277 | peptide binding(GO:0042277) |
| 0.0 | 0.1 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.1 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 1.8 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.5 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 2.0 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |