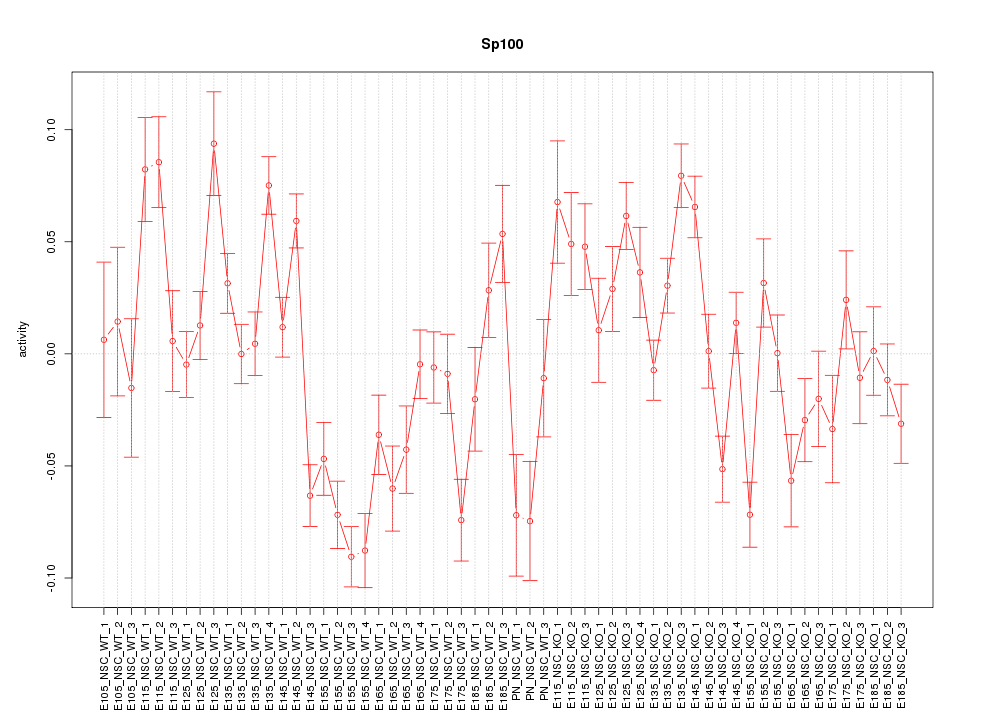
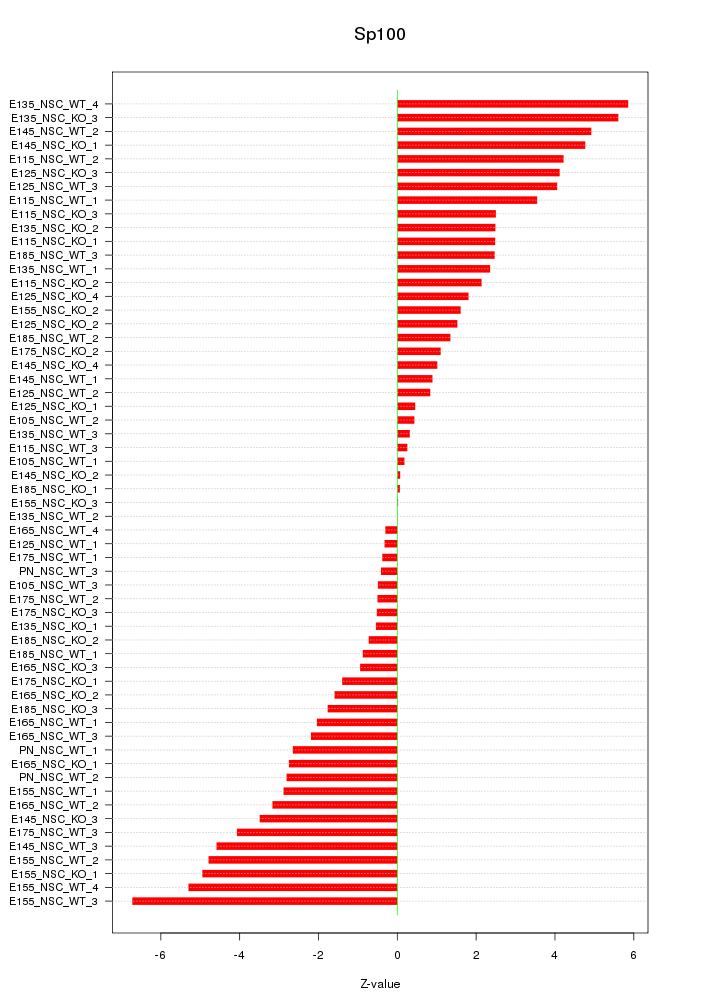
Motif ID: Sp100
Z-value: 2.784

Transcription factors associated with Sp100:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Sp100 | ENSMUSG00000026222.10 | Sp100 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sp100 | mm10_v2_chr1_+_85650044_85650052 | 0.31 | 1.6e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 8.0 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 1.5 | 7.6 | GO:0021764 | amygdala development(GO:0021764) |
| 1.5 | 10.4 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 1.4 | 8.2 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 1.3 | 6.6 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 1.2 | 3.5 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 1.1 | 3.2 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 1.1 | 10.5 | GO:0046036 | GTP biosynthetic process(GO:0006183) CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 1.0 | 4.9 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.9 | 6.9 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.9 | 2.6 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.8 | 2.4 | GO:2000852 | regulation of corticosterone secretion(GO:2000852) |
| 0.8 | 2.3 | GO:0060126 | somatotropin secreting cell differentiation(GO:0060126) |
| 0.8 | 2.3 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 0.7 | 12.7 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.7 | 2.1 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.6 | 1.7 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.6 | 2.9 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.6 | 2.3 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.5 | 2.1 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.5 | 2.6 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.5 | 4.5 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.5 | 3.4 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.5 | 9.2 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.5 | 5.1 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.4 | 1.3 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.4 | 2.8 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.4 | 2.3 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.4 | 1.1 | GO:0032240 | negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.3 | 2.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.3 | 3.2 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.3 | 5.4 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.3 | 1.4 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.2 | 3.0 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.2 | 4.3 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.2 | 1.2 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) |
| 0.2 | 4.6 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.2 | 2.3 | GO:0006273 | lagging strand elongation(GO:0006273) |
| 0.2 | 1.6 | GO:0090209 | negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.2 | 2.4 | GO:1990173 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.2 | 7.7 | GO:0021511 | spinal cord patterning(GO:0021511) |
| 0.2 | 1.9 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.2 | 5.0 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.2 | 3.6 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.2 | 16.4 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.2 | 1.1 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.2 | 2.8 | GO:0036065 | fucosylation(GO:0036065) |
| 0.2 | 0.7 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.2 | 2.0 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.2 | 2.5 | GO:0045910 | negative regulation of DNA recombination(GO:0045910) |
| 0.2 | 3.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.2 | 1.6 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.2 | 0.3 | GO:0040038 | polar body extrusion after meiotic divisions(GO:0040038) |
| 0.2 | 1.5 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 0.4 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.1 | 1.3 | GO:0045191 | regulation of isotype switching(GO:0045191) |
| 0.1 | 1.2 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.1 | 2.7 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 | 1.8 | GO:0019363 | pyridine nucleotide biosynthetic process(GO:0019363) |
| 0.1 | 1.3 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 3.4 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.1 | 2.3 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.1 | 1.1 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.1 | 2.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 1.4 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.1 | 2.3 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.1 | 1.9 | GO:1903861 | regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 3.8 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.1 | 0.8 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 10.2 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.1 | 2.5 | GO:2000279 | negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.1 | 1.4 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 1.3 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 1.0 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 1.9 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.1 | 4.1 | GO:0007338 | single fertilization(GO:0007338) |
| 0.1 | 3.8 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.1 | 3.7 | GO:0070613 | regulation of protein processing(GO:0070613) |
| 0.0 | 1.9 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 1.8 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 1.0 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.0 | 2.8 | GO:0034644 | cellular response to UV(GO:0034644) |
| 0.0 | 2.6 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 2.5 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.4 | GO:0061000 | negative regulation of dendritic spine development(GO:0061000) |
| 0.0 | 0.5 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.0 | 2.7 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 8.7 | GO:0007059 | chromosome segregation(GO:0007059) |
| 0.0 | 0.8 | GO:0006625 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 1.9 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 1.9 | GO:0000077 | DNA damage checkpoint(GO:0000077) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 2.2 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.4 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 1.0 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 2.3 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.0 | 0.8 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 1.3 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.0 | 1.3 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 2.1 | GO:0098792 | positive regulation of defense response to virus by host(GO:0002230) xenophagy(GO:0098792) |
| 0.0 | 0.8 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.0 | 0.3 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.8 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.0 | 0.5 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 1.1 | GO:0051168 | nuclear export(GO:0051168) |
| 0.0 | 0.3 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.6 | GO:0043293 | apoptosome(GO:0043293) |
| 0.9 | 8.0 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.9 | 5.2 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.8 | 2.4 | GO:0031417 | NatC complex(GO:0031417) |
| 0.8 | 2.3 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.8 | 2.3 | GO:1990047 | spindle matrix(GO:1990047) |
| 0.7 | 2.1 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.6 | 6.3 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.5 | 5.1 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.5 | 6.6 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.4 | 9.4 | GO:0001741 | XY body(GO:0001741) |
| 0.4 | 2.5 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.3 | 10.3 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.3 | 2.3 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.2 | 3.9 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 0.2 | 2.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.2 | 10.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 1.1 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.2 | 2.6 | GO:0048188 | MLL3/4 complex(GO:0044666) Set1C/COMPASS complex(GO:0048188) |
| 0.2 | 3.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 7.6 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.2 | 1.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.2 | 3.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.2 | 2.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 2.7 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.1 | 1.3 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 10.2 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.1 | 8.2 | GO:0005657 | replication fork(GO:0005657) |
| 0.1 | 1.7 | GO:0001527 | microfibril(GO:0001527) |
| 0.1 | 1.4 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 2.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 1.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 6.5 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 0.9 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 7.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 0.4 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.1 | 7.3 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 6.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 1.5 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 4.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 0.5 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.1 | 2.6 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 1.3 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 1.7 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.1 | 1.2 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 0.3 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 5.0 | GO:0000313 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.1 | 1.9 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 1.3 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 2.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.6 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 1.2 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 1.9 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 1.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 2.4 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 1.8 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 265.8 | GO:0005575 | cellular_component(GO:0005575) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.2 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 2.0 | 8.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 1.2 | 3.5 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 1.1 | 3.2 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.9 | 3.7 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.9 | 6.9 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.7 | 5.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.6 | 10.4 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.6 | 2.4 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.5 | 10.5 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.5 | 2.5 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.5 | 5.0 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.4 | 2.3 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.4 | 1.1 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.4 | 2.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.3 | 2.3 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.3 | 1.0 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.3 | 2.2 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.3 | 4.9 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.3 | 3.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.2 | 2.6 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.2 | 1.8 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.2 | 9.5 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.2 | 1.6 | GO:0046790 | virion binding(GO:0046790) |
| 0.2 | 8.1 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.2 | 6.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 4.3 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.2 | 1.3 | GO:0018741 | alkyl sulfatase activity(GO:0018741) endosulfan hemisulfate sulfatase activity(GO:0034889) endosulfan sulfate hydrolase activity(GO:0034902) |
| 0.2 | 7.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.2 | 3.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.2 | 1.4 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.2 | 2.7 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.1 | 1.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 1.9 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 2.5 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 7.6 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 2.1 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 2.0 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 3.5 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 1.3 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.1 | 2.7 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 1.7 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.1 | 1.8 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.5 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 2.3 | GO:0050694 | galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 1.0 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 1.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 4.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 1.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 2.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 2.0 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.7 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 1.6 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 12.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 4.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 2.2 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 2.4 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 10.3 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 1.1 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.3 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 1.1 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 8.4 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 2.8 | GO:0035496 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) O antigen polymerase activity(GO:0008755) lipopolysaccharide-1,6-galactosyltransferase activity(GO:0008921) cellulose synthase activity(GO:0016759) 9-phenanthrol UDP-glucuronosyltransferase activity(GO:0018715) 1-phenanthrol glycosyltransferase activity(GO:0018716) 9-phenanthrol glycosyltransferase activity(GO:0018717) 1,2-dihydroxy-phenanthrene glycosyltransferase activity(GO:0018718) phenanthrol glycosyltransferase activity(GO:0019112) alpha-1,2-galactosyltransferase activity(GO:0031278) dolichyl pyrophosphate Man7GlcNAc2 alpha-1,3-glucosyltransferase activity(GO:0033556) endogalactosaminidase activity(GO:0033931) lipopolysaccharide-1,5-galactosyltransferase activity(GO:0035496) dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase activity(GO:0042283) inositol phosphoceramide synthase activity(GO:0045140) alpha-(1->3)-fucosyltransferase activity(GO:0046920) indole-3-butyrate beta-glucosyltransferase activity(GO:0052638) salicylic acid glucosyltransferase (ester-forming) activity(GO:0052639) salicylic acid glucosyltransferase (glucoside-forming) activity(GO:0052640) benzoic acid glucosyltransferase activity(GO:0052641) chondroitin hydrolase activity(GO:0052757) dolichyl-pyrophosphate Man7GlcNAc2 alpha-1,6-mannosyltransferase activity(GO:0052824) cytokinin 9-beta-glucosyltransferase activity(GO:0080062) |
| 0.0 | 1.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.8 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 1.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 2.6 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 2.3 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.0 | 0.4 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.0 | 0.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 4.9 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 1.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 1.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.5 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.6 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 1.2 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 1.9 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.6 | GO:0044824 | integrase activity(GO:0008907) T/G mismatch-specific endonuclease activity(GO:0043765) retroviral integrase activity(GO:0044823) retroviral 3' processing activity(GO:0044824) |
| 0.0 | 210.8 | GO:0003674 | molecular_function(GO:0003674) |