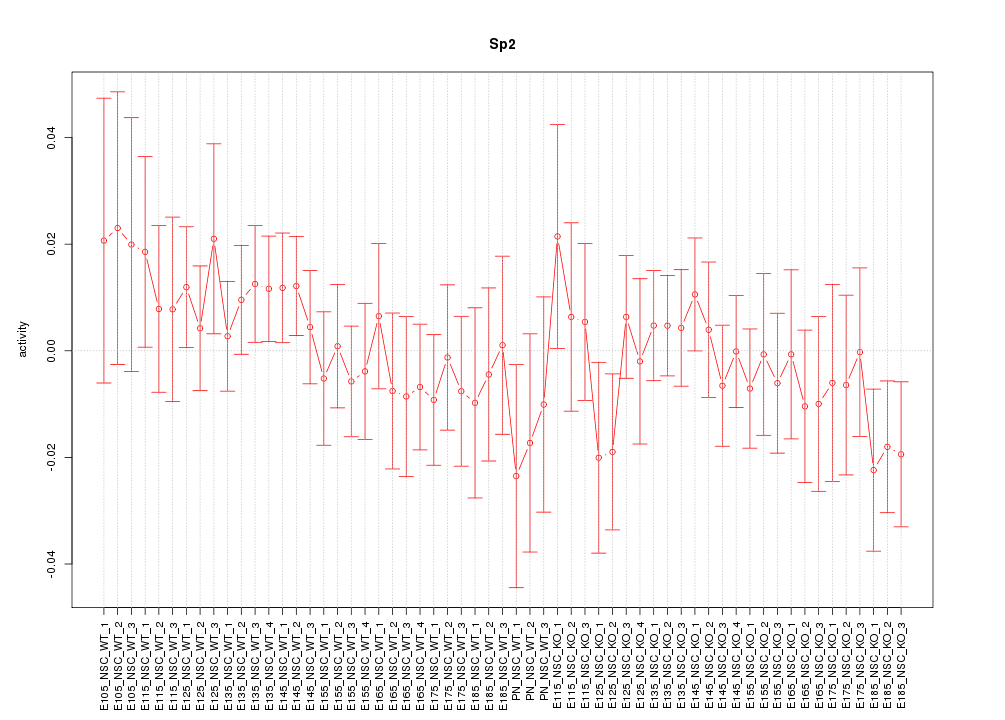
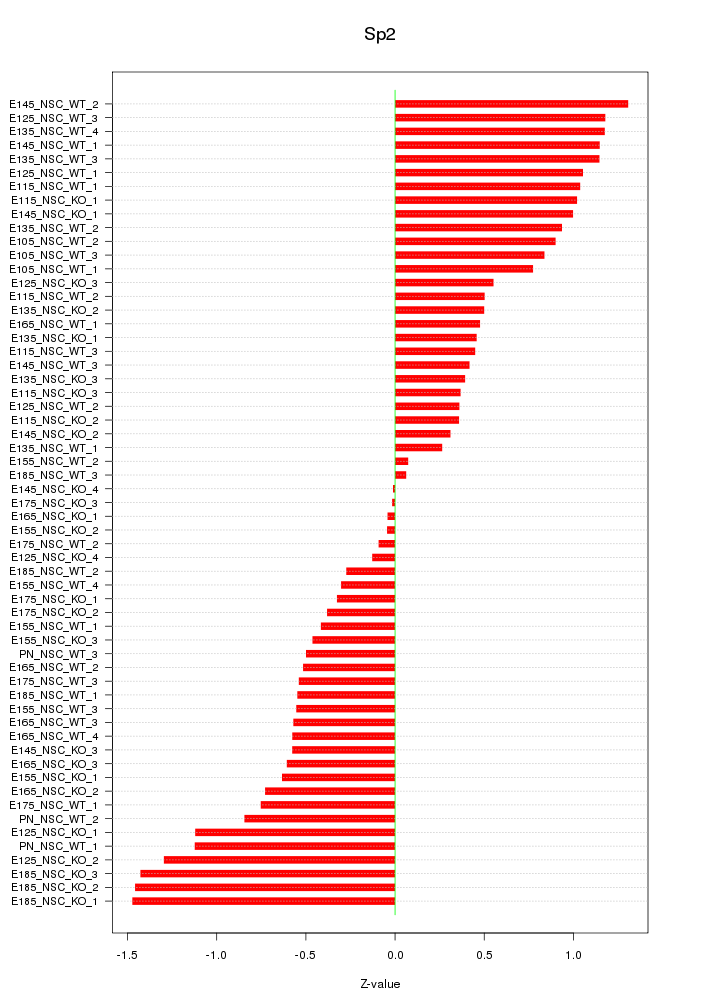
Motif ID: Sp2
Z-value: 0.748
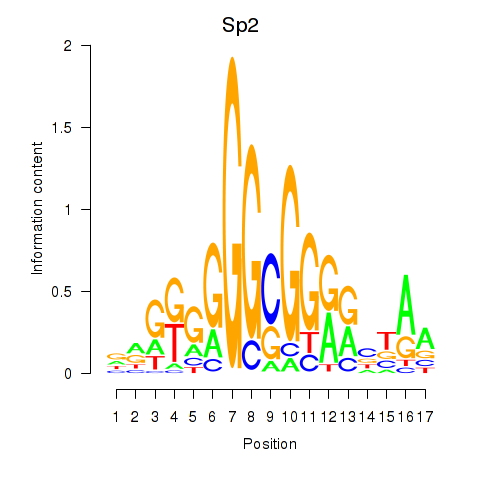
Transcription factors associated with Sp2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Sp2 | ENSMUSG00000018678.6 | Sp2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sp2 | mm10_v2_chr11_-_96968655_96968673 | -0.09 | 5.2e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 1.1 | 14.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 1.0 | 3.1 | GO:0000087 | mitotic M phase(GO:0000087) |
| 0.9 | 2.8 | GO:0097402 | neuroblast migration(GO:0097402) |
| 0.8 | 2.5 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.8 | 2.4 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.8 | 3.0 | GO:0035795 | negative regulation of mitochondrial membrane permeability(GO:0035795) |
| 0.7 | 2.0 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.7 | 2.0 | GO:1901228 | positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.6 | 2.6 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.6 | 1.9 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.6 | 8.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.6 | 2.9 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.6 | 1.7 | GO:0001193 | maintenance of transcriptional fidelity during DNA-templated transcription elongation(GO:0001192) maintenance of transcriptional fidelity during DNA-templated transcription elongation from RNA polymerase II promoter(GO:0001193) |
| 0.5 | 1.1 | GO:0090289 | regulation of osteoclast proliferation(GO:0090289) |
| 0.5 | 3.8 | GO:0031652 | positive regulation of heat generation(GO:0031652) |
| 0.5 | 1.6 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.5 | 1.5 | GO:0009153 | purine deoxyribonucleotide biosynthetic process(GO:0009153) |
| 0.5 | 1.4 | GO:0010958 | regulation of amino acid import(GO:0010958) regulation of L-arginine import(GO:0010963) |
| 0.4 | 2.7 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.4 | 1.3 | GO:0046370 | fructose biosynthetic process(GO:0046370) |
| 0.4 | 2.7 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.4 | 1.3 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.4 | 1.7 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.4 | 1.2 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.4 | 1.5 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) negative regulation of respiratory burst(GO:0060268) |
| 0.4 | 1.5 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.4 | 3.8 | GO:1901030 | positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901030) |
| 0.4 | 1.1 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.4 | 1.5 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.4 | 1.4 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.4 | 1.4 | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) |
| 0.3 | 1.0 | GO:0045472 | response to ether(GO:0045472) |
| 0.3 | 2.4 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.3 | 2.7 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.3 | 1.7 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.3 | 2.7 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.3 | 1.6 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.3 | 1.3 | GO:0002741 | positive regulation of cytokine secretion involved in immune response(GO:0002741) |
| 0.3 | 1.3 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.3 | 3.8 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.3 | 0.9 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.3 | 2.1 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.3 | 1.2 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.3 | 2.1 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.3 | 1.7 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.3 | 0.8 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.3 | 0.8 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.3 | 0.8 | GO:0090526 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.3 | 1.3 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.3 | 0.3 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) |
| 0.3 | 4.6 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.3 | 1.3 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.2 | 1.2 | GO:1902856 | negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.2 | 2.9 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.2 | 0.7 | GO:0070844 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.2 | 0.9 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.2 | 1.4 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.2 | 1.9 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.2 | 1.0 | GO:1902309 | positive regulation of mast cell chemotaxis(GO:0060754) negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.2 | 1.7 | GO:0015074 | DNA integration(GO:0015074) |
| 0.2 | 0.6 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 0.8 | GO:0071600 | otic vesicle morphogenesis(GO:0071600) |
| 0.2 | 1.2 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.2 | 0.6 | GO:0070384 | Harderian gland development(GO:0070384) |
| 0.2 | 0.8 | GO:0072592 | oxygen metabolic process(GO:0072592) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.2 | 0.8 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.2 | 0.9 | GO:0043951 | negative regulation of phospholipase activity(GO:0010519) negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.2 | 1.3 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.2 | 0.9 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.2 | 0.9 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.2 | 1.6 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.2 | 0.7 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.2 | 1.6 | GO:0042538 | hyperosmotic salinity response(GO:0042538) |
| 0.2 | 0.9 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.2 | 2.6 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.2 | 0.7 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.2 | 2.3 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.2 | 0.3 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.2 | 0.5 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.2 | 2.1 | GO:0070445 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.2 | 1.6 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.2 | 1.9 | GO:0070633 | transepithelial transport(GO:0070633) |
| 0.2 | 0.6 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.2 | 0.6 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.2 | 0.8 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.1 | 3.6 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 0.6 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 0.7 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.1 | 1.3 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.1 | 1.0 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 | 1.2 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 1.4 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.1 | 2.0 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.7 | GO:0003273 | cell migration involved in endocardial cushion formation(GO:0003273) |
| 0.1 | 0.6 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 0.1 | 0.6 | GO:1904690 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.1 | 1.2 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.1 | 1.3 | GO:0000059 | protein import into nucleus, docking(GO:0000059) |
| 0.1 | 1.6 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 | 0.2 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.1 | 0.9 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 | 1.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 0.6 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.8 | GO:0021747 | cochlear nucleus development(GO:0021747) |
| 0.1 | 0.8 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 2.5 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.1 | 0.7 | GO:0070458 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.1 | 0.3 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.3 | GO:0051383 | kinetochore organization(GO:0051383) |
| 0.1 | 1.7 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 3.6 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.1 | 4.7 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.1 | 1.3 | GO:2000010 | positive regulation of protein localization to cell surface(GO:2000010) |
| 0.1 | 3.2 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 0.7 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.1 | 1.0 | GO:2000059 | negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.1 | 0.3 | GO:1902915 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 | 0.3 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 1.1 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.7 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 1.7 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.4 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.4 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 1.6 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.3 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.1 | 0.4 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.1 | 0.8 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.3 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 2.7 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 0.3 | GO:0046476 | glycosylceramide biosynthetic process(GO:0046476) |
| 0.1 | 0.2 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.1 | 0.6 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.2 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 | 1.1 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 2.1 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.1 | 0.3 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.1 | 2.0 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.1 | 0.5 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.1 | 0.3 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.1 | 0.2 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.1 | 0.5 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.1 | 0.1 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.1 | 0.5 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.1 | 0.4 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.4 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 0.4 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 1.2 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 0.4 | GO:0007135 | meiosis II(GO:0007135) |
| 0.1 | 0.3 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.1 | 0.7 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 0.8 | GO:0031442 | positive regulation of mRNA 3'-end processing(GO:0031442) |
| 0.1 | 0.4 | GO:0001771 | immunological synapse formation(GO:0001771) |
| 0.1 | 0.2 | GO:0032471 | negative regulation of endoplasmic reticulum calcium ion concentration(GO:0032471) |
| 0.1 | 1.2 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 1.2 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.1 | 0.9 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.1 | 0.5 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.1 | 0.8 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.1 | 0.7 | GO:0046697 | decidualization(GO:0046697) |
| 0.1 | 0.6 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 3.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.8 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.3 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 1.3 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.0 | 0.8 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 0.0 | 0.4 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.3 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.2 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.0 | 0.1 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.0 | 0.7 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.4 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.0 | 0.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 2.9 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.6 | GO:0051220 | cytoplasmic sequestering of protein(GO:0051220) |
| 0.0 | 1.1 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.7 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.5 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 1.2 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.1 | GO:1904749 | protein localization to nucleolus(GO:1902570) regulation of protein localization to nucleolus(GO:1904749) positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.0 | 1.1 | GO:0006084 | acetyl-CoA metabolic process(GO:0006084) |
| 0.0 | 0.2 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.6 | GO:0010388 | cullin deneddylation(GO:0010388) |
| 0.0 | 0.7 | GO:0060561 | apoptotic process involved in morphogenesis(GO:0060561) |
| 0.0 | 0.1 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.5 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.2 | GO:0021559 | trigeminal nerve development(GO:0021559) |
| 0.0 | 0.7 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.6 | GO:0005980 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.5 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.6 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.1 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 1.5 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.2 | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway(GO:2001243) |
| 0.0 | 0.2 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.2 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.8 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 1.0 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.2 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0048382 | neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) mesendoderm development(GO:0048382) |
| 0.0 | 0.3 | GO:0051352 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) |
| 0.0 | 0.8 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.5 | GO:0007187 | G-protein coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger(GO:0007187) |
| 0.0 | 0.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.8 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.2 | GO:1901409 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.7 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.7 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.0 | 0.8 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.4 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.5 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 0.1 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 2.6 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.0 | 1.2 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 0.9 | GO:0000077 | DNA damage checkpoint(GO:0000077) |
| 0.0 | 0.3 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.8 | GO:0051782 | negative regulation of cell division(GO:0051782) |
| 0.0 | 0.1 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 1.2 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0042167 | heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.3 | GO:0048741 | skeletal muscle fiber development(GO:0048741) |
| 0.0 | 1.2 | GO:0032434 | regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032434) |
| 0.0 | 0.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.6 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.6 | GO:0031123 | RNA 3'-end processing(GO:0031123) |
| 0.0 | 0.3 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.2 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.6 | 2.4 | GO:0090537 | CERF complex(GO:0090537) |
| 0.4 | 2.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.4 | 1.2 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.4 | 2.4 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.3 | 2.0 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.3 | 2.6 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.3 | 1.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.3 | 0.9 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.3 | 1.4 | GO:0034715 | U7 snRNP(GO:0005683) pICln-Sm protein complex(GO:0034715) |
| 0.3 | 1.9 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.3 | 1.3 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.2 | 0.9 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.2 | 0.7 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.2 | 1.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.2 | 0.6 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.2 | 2.2 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 0.2 | 0.8 | GO:0097413 | Lewy body(GO:0097413) |
| 0.2 | 1.7 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.2 | 2.0 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.2 | 2.7 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.2 | 1.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 0.9 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 0.6 | GO:0031230 | intrinsic component of cell outer membrane(GO:0031230) integral component of cell outer membrane(GO:0045203) |
| 0.1 | 1.7 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 1.8 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 2.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 2.6 | GO:0045120 | pronucleus(GO:0045120) |
| 0.1 | 0.2 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.1 | 1.2 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 1.5 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.1 | 0.6 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 4.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 2.3 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 1.2 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 0.3 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.3 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.1 | 0.8 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 0.8 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.1 | 1.1 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 0.6 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.5 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.1 | 2.6 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 0.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 1.1 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.1 | 4.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 1.0 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.2 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 0.5 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.1 | 1.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.3 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 1.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 0.6 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 0.4 | GO:0090543 | Flemming body(GO:0090543) |
| 0.1 | 1.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.8 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 0.7 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 1.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 1.3 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 0.4 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.7 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.8 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 1.7 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 4.2 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.9 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.4 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 1.1 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.3 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.5 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 2.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 4.9 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.2 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 2.5 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.9 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 1.2 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 2.0 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.7 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.9 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.3 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 3.5 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.9 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 2.6 | GO:0031513 | nonmotile primary cilium(GO:0031513) |
| 0.0 | 0.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.4 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.4 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 1.9 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.6 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.1 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 1.6 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.5 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 28.9 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 0.8 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 1.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.6 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 13.1 | GO:0019841 | retinol binding(GO:0019841) |
| 1.6 | 8.2 | GO:0018121 | imidazoleglycerol-phosphate synthase activity(GO:0000107) NAD(P)-cysteine ADP-ribosyltransferase activity(GO:0018071) NAD(P)-asparagine ADP-ribosyltransferase activity(GO:0018121) NAD(P)-serine ADP-ribosyltransferase activity(GO:0018127) 7-cyano-7-deazaguanine tRNA-ribosyltransferase activity(GO:0043867) purine deoxyribosyltransferase activity(GO:0044102) |
| 0.8 | 2.4 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.6 | 3.0 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.6 | 2.9 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.5 | 2.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.5 | 1.6 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.5 | 1.4 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) U7 snRNA binding(GO:0071209) |
| 0.5 | 1.9 | GO:0003681 | bent DNA binding(GO:0003681) |
| 0.4 | 3.5 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.4 | 1.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.4 | 1.2 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.4 | 1.1 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.4 | 1.1 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.3 | 2.6 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.3 | 0.9 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.3 | 1.2 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.3 | 2.0 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.3 | 1.3 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.3 | 0.8 | GO:0005118 | sevenless binding(GO:0005118) |
| 0.3 | 0.8 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.3 | 0.8 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.2 | 0.9 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.2 | 1.4 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.2 | 1.4 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.2 | 2.5 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.2 | 1.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.2 | 1.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.2 | 1.0 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 0.8 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.2 | 3.8 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.2 | 1.5 | GO:0103116 | alpha-D-galactofuranose transporter activity(GO:0103116) steroid hormone binding(GO:1990239) |
| 0.2 | 3.8 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.2 | 0.5 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.2 | 0.7 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.2 | 1.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.2 | 1.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.2 | 0.3 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.2 | 0.6 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.2 | 2.5 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 2.8 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.4 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 1.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 3.0 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 0.8 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 1.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 6.2 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.1 | 0.6 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 1.0 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 0.8 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 4.7 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 1.7 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 1.7 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.6 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.1 | 1.5 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 1.1 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.1 | 0.6 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.1 | 1.7 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 0.6 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.1 | 0.6 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 1.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.5 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.1 | 0.7 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.1 | 0.3 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.6 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 0.7 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.1 | 0.9 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 1.6 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 0.9 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 3.4 | GO:0018602 | sulfonate dioxygenase activity(GO:0000907) 2,4-dichlorophenoxyacetate alpha-ketoglutarate dioxygenase activity(GO:0018602) hypophosphite dioxygenase activity(GO:0034792) gibberellin 2-beta-dioxygenase activity(GO:0045543) C-19 gibberellin 2-beta-dioxygenase activity(GO:0052634) C-20 gibberellin 2-beta-dioxygenase activity(GO:0052635) |
| 0.1 | 0.3 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.1 | 0.7 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) |
| 0.1 | 1.2 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 1.7 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.1 | 1.1 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 0.6 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.8 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.5 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.1 | 0.6 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.1 | 1.5 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.1 | 0.3 | GO:0004921 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.1 | 1.0 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.7 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 1.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 4.2 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 1.2 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 1.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.2 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 1.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 2.2 | GO:0016749 | N-succinyltransferase activity(GO:0016749) |
| 0.1 | 1.7 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.1 | 6.9 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 1.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.5 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 1.7 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 0.6 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 3.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.9 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.5 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 1.1 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.3 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 11.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.4 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 1.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.4 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.4 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 1.9 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.9 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.6 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.2 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.9 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.6 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:0019145 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.1 | GO:0048045 | 4-hydroxybenzoate octaprenyltransferase activity(GO:0008412) protoheme IX farnesyltransferase activity(GO:0008495) (S)-2,3-di-O-geranylgeranylglyceryl phosphate synthase activity(GO:0043888) cadaverine aminopropyltransferase activity(GO:0043918) agmatine aminopropyltransferase activity(GO:0043919) 1,4-dihydroxy-2-naphthoate octaprenyltransferase activity(GO:0046428) trans-pentaprenyltranstransferase activity(GO:0048045) ATP dimethylallyltransferase activity(GO:0052622) ADP dimethylallyltransferase activity(GO:0052623) |
| 0.0 | 0.7 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.1 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 2.0 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 1.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) |
| 0.0 | 1.1 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 1.1 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.5 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.3 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.2 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.0 | 0.8 | GO:0045502 | dynein binding(GO:0045502) |
| 0.0 | 0.4 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.1 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.0 | 0.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.0 | 1.5 | GO:0001076 | transcription factor activity, RNA polymerase II transcription factor binding(GO:0001076) |
| 0.0 | 2.5 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.3 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.0 | 0.8 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 1.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.2 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.0 | 0.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.3 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0034951 | pivalyl-CoA mutase activity(GO:0034784) o-hydroxylaminobenzoate mutase activity(GO:0034951) lupeol synthase activity(GO:0042299) beta-amyrin synthase activity(GO:0042300) baruol synthase activity(GO:0080011) |
| 0.0 | 0.2 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.1 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) poly(G) binding(GO:0034046) |
| 0.0 | 0.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 7.5 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.7 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.8 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 1.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.7 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.6 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.4 | GO:0000049 | tRNA binding(GO:0000049) |