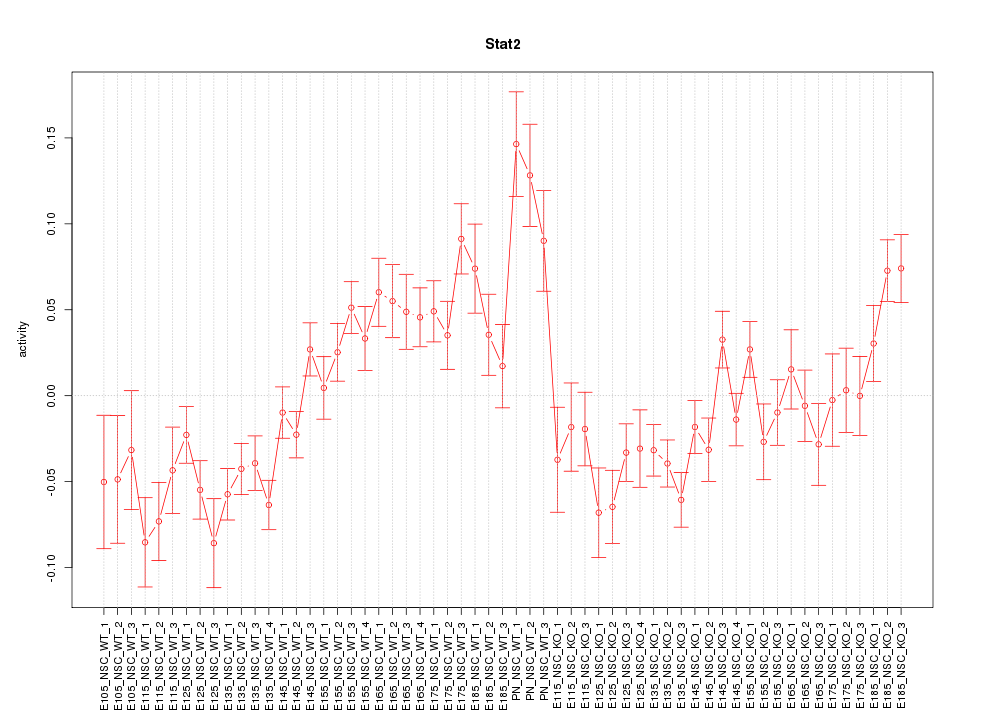
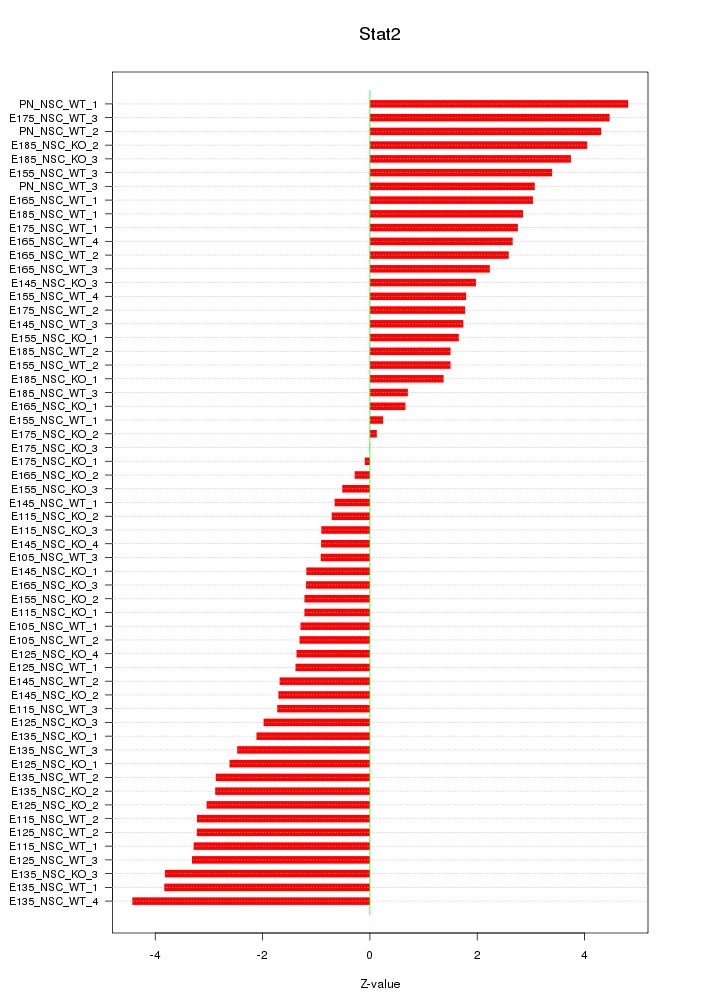
Motif ID: Stat2
Z-value: 2.416

Transcription factors associated with Stat2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Stat2 | ENSMUSG00000040033.9 | Stat2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Stat2 | mm10_v2_chr10_+_128270546_128270577 | 0.64 | 4.6e-08 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.7 | 32.1 | GO:0042560 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 6.9 | 20.7 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 6.7 | 33.3 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 5.5 | 16.6 | GO:1902524 | negative regulation of interferon-alpha production(GO:0032687) interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) negative regulation of interferon-beta biosynthetic process(GO:0045358) positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 4.7 | 14.0 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) metanephric glomerular mesangium development(GO:0072223) metanephric glomerular mesangial cell proliferation involved in metanephros development(GO:0072262) |
| 4.4 | 13.3 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 3.6 | 14.5 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 2.9 | 11.6 | GO:0009597 | detection of virus(GO:0009597) |
| 2.8 | 8.5 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 2.6 | 7.7 | GO:0046077 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 2.6 | 7.7 | GO:0006533 | aspartate catabolic process(GO:0006533) D-amino acid catabolic process(GO:0019478) |
| 2.6 | 20.4 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 2.4 | 16.8 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 2.4 | 37.9 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 2.3 | 6.9 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 2.1 | 14.8 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 2.1 | 39.0 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 2.0 | 12.1 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 2.0 | 7.8 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 1.8 | 12.8 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 1.7 | 8.7 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 1.6 | 4.7 | GO:0002578 | negative regulation of antigen processing and presentation(GO:0002578) |
| 1.4 | 7.0 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 1.3 | 38.9 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 1.3 | 3.8 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 1.3 | 8.9 | GO:0015840 | urea transport(GO:0015840) urea transmembrane transport(GO:0071918) |
| 1.3 | 5.1 | GO:0032439 | endosome localization(GO:0032439) |
| 1.2 | 12.4 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 1.2 | 11.8 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 1.2 | 16.2 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 1.0 | 4.0 | GO:0032512 | regulation of protein phosphatase type 2B activity(GO:0032512) positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 1.0 | 27.7 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.9 | 8.4 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.9 | 4.3 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 0.8 | 4.1 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.8 | 5.5 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.8 | 4.5 | GO:1903056 | regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.7 | 5.2 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.7 | 2.1 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.7 | 5.5 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.6 | 9.7 | GO:0034340 | response to type I interferon(GO:0034340) |
| 0.6 | 1.9 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.6 | 2.4 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.6 | 1.8 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 0.6 | 2.3 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.6 | 1.7 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.6 | 8.5 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.6 | 4.4 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.5 | 1.6 | GO:0048611 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.5 | 3.2 | GO:0035989 | tendon development(GO:0035989) |
| 0.5 | 25.0 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.5 | 20.8 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.5 | 4.2 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.5 | 4.5 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.4 | 2.6 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.4 | 10.7 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.4 | 3.4 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.4 | 5.8 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.4 | 1.2 | GO:0050713 | negative regulation of interleukin-1 secretion(GO:0050711) negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.4 | 3.8 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.3 | 2.8 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.3 | 2.7 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.3 | 2.5 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.3 | 3.5 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.2 | 4.1 | GO:0032148 | activation of protein kinase B activity(GO:0032148) |
| 0.2 | 0.7 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.2 | 4.2 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.2 | 1.4 | GO:0051204 | protein insertion into mitochondrial membrane(GO:0051204) |
| 0.2 | 4.7 | GO:0001783 | B cell apoptotic process(GO:0001783) |
| 0.2 | 1.2 | GO:0051036 | regulation of endosome size(GO:0051036) |
| 0.2 | 0.8 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 0.4 | GO:0070944 | neutrophil mediated killing of bacterium(GO:0070944) |
| 0.1 | 0.7 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.1 | 4.1 | GO:0015991 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.1 | 5.3 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 0.3 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.1 | 0.7 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.1 | 1.6 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 0.3 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.1 | 0.2 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.1 | 9.1 | GO:0019395 | fatty acid oxidation(GO:0019395) |
| 0.1 | 5.0 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.1 | 2.4 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.1 | 2.5 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.1 | 2.1 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.1 | 1.1 | GO:0009072 | aromatic amino acid family metabolic process(GO:0009072) |
| 0.1 | 1.4 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.1 | 11.4 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.1 | 4.6 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.1 | 0.2 | GO:0019085 | early viral transcription(GO:0019085) late viral transcription(GO:0019086) |
| 0.0 | 2.7 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.0 | 3.4 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.9 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 1.9 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 5.4 | GO:0043087 | regulation of GTPase activity(GO:0043087) |
| 0.0 | 5.8 | GO:0060047 | heart contraction(GO:0060047) |
| 0.0 | 2.6 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 5.0 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 3.9 | GO:0042384 | cilium assembly(GO:0042384) |
| 0.0 | 1.6 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 6.6 | GO:0006914 | autophagy(GO:0006914) |
| 0.0 | 0.1 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 1.0 | GO:0045727 | positive regulation of translation(GO:0045727) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 11.1 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 1.9 | 20.8 | GO:0043083 | synaptic cleft(GO:0043083) |
| 1.5 | 10.7 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 1.5 | 6.0 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 1.4 | 5.8 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 1.2 | 13.7 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 1.1 | 4.5 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 1.1 | 12.4 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.8 | 3.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.8 | 33.1 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.7 | 3.7 | GO:1990357 | terminal web(GO:1990357) |
| 0.7 | 5.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.6 | 5.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.5 | 5.1 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.5 | 8.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.4 | 39.1 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.4 | 6.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.4 | 3.4 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.3 | 1.6 | GO:1990745 | EARP complex(GO:1990745) |
| 0.3 | 2.7 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.3 | 14.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.3 | 4.0 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.2 | 88.4 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.2 | 1.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.2 | 7.0 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.2 | 1.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.2 | 0.7 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.2 | 4.2 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.2 | 2.1 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 4.0 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.2 | 2.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 11.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 0.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 33.3 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.1 | 2.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 2.1 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 1.4 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 2.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 7.7 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 8.0 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 7.7 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.1 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 2.2 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 22.1 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.9 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 38.4 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 6.1 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.9 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 13.0 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 71.6 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 2.0 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 9.6 | GO:0005829 | cytosol(GO:0005829) |
| 0.0 | 1.0 | GO:0045211 | postsynaptic membrane(GO:0045211) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.7 | 32.1 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 8.3 | 33.3 | GO:0038049 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 3.0 | 12.1 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 3.0 | 20.8 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 2.9 | 37.9 | GO:0016443 | bidentate ribonuclease III activity(GO:0016443) |
| 2.8 | 14.0 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 2.6 | 7.7 | GO:0004127 | cytidylate kinase activity(GO:0004127) uridylate kinase activity(GO:0009041) |
| 2.1 | 8.4 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 2.0 | 6.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 1.9 | 7.7 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) aspartate oxidase activity(GO:0015922) |
| 1.9 | 7.5 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 1.6 | 14.8 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 1.6 | 20.4 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 1.5 | 8.9 | GO:0015265 | urea channel activity(GO:0015265) |
| 1.4 | 33.1 | GO:0005521 | lamin binding(GO:0005521) |
| 1.4 | 4.2 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 1.4 | 6.9 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 1.3 | 3.8 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 1.1 | 4.6 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 1.1 | 5.3 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 1.0 | 9.9 | GO:0052872 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) phenanthrene 9,10-monooxygenase activity(GO:0018636) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) tocotrienol omega-hydroxylase activity(GO:0052872) thalianol hydroxylase activity(GO:0080014) |
| 1.0 | 12.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.9 | 12.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.9 | 18.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.9 | 5.1 | GO:0046978 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.7 | 3.5 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.6 | 1.9 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.6 | 5.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.6 | 6.2 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.6 | 4.2 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.6 | 8.5 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.5 | 2.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.5 | 12.0 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.5 | 1.5 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.5 | 4.0 | GO:0043495 | protein anchor(GO:0043495) |
| 0.5 | 4.2 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.5 | 3.7 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.4 | 3.4 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.4 | 4.0 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.4 | 1.5 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.3 | 11.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.3 | 5.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.3 | 2.2 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled adenosine receptor activity(GO:0001609) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.3 | 0.9 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.3 | 43.3 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.3 | 11.4 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.2 | 2.4 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.2 | 15.8 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.2 | 4.1 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.2 | 1.6 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.2 | 4.6 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.2 | 13.7 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.2 | 2.7 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.2 | 13.4 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.2 | 0.8 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.2 | 2.1 | GO:0031386 | protein tag(GO:0031386) |
| 0.2 | 4.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 4.1 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 4.5 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 8.0 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.1 | 0.3 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.1 | 11.1 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.1 | 1.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.9 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 1.2 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.1 | 1.2 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 10.7 | GO:0003729 | mRNA binding(GO:0003729) |
| 0.1 | 33.3 | GO:0005525 | GTP binding(GO:0005525) |
| 0.1 | 24.4 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 4.1 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 2.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 2.8 | GO:0004120 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) photoreceptor cyclic-nucleotide phosphodiesterase activity(GO:0004120) 7,8-dihydro-D-neopterin 2',3'-cyclic phosphate phosphodiesterase activity(GO:0044688) inositol phosphosphingolipid phospholipase activity(GO:0052712) inositol phosphorylceramide phospholipase activity(GO:0052713) mannosyl-inositol phosphorylceramide phospholipase activity(GO:0052714) mannosyl-diinositol phosphorylceramide phospholipase activity(GO:0052715) |
| 0.1 | 2.1 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 9.8 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 0.7 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.3 | GO:0000982 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) |
| 0.0 | 5.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 5.1 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 3.5 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 7.1 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 1.6 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 1.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 8.5 | GO:0016887 | ATPase activity(GO:0016887) |
| 0.0 | 0.1 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.0 | 1.8 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 1.5 | GO:0043851 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) rRNA (uridine-2'-O-)-methyltransferase activity(GO:0008650) rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) selenocysteine methyltransferase activity(GO:0016205) rRNA (adenine) methyltransferase activity(GO:0016433) rRNA (cytosine) methyltransferase activity(GO:0016434) rRNA (guanine) methyltransferase activity(GO:0016435) 1-phenanthrol methyltransferase activity(GO:0018707) protein-arginine N5-methyltransferase activity(GO:0019702) dimethylarsinite methyltransferase activity(GO:0034541) 4,5-dihydroxybenzo(a)pyrene methyltransferase activity(GO:0034807) 1-hydroxypyrene methyltransferase activity(GO:0034931) 1-hydroxy-6-methoxypyrene methyltransferase activity(GO:0034933) demethylmenaquinone methyltransferase activity(GO:0043770) cobalt-precorrin-6B C5-methyltransferase activity(GO:0043776) cobalt-precorrin-7 C15-methyltransferase activity(GO:0043777) cobalt-precorrin-5B C1-methyltransferase activity(GO:0043780) cobalt-precorrin-3 C17-methyltransferase activity(GO:0043782) dimethylamine methyltransferase activity(GO:0043791) hydroxyneurosporene-O-methyltransferase activity(GO:0043803) tRNA (adenine-57, 58-N(1)-) methyltransferase activity(GO:0043827) methylamine-specific methylcobalamin:coenzyme M methyltransferase activity(GO:0043833) trimethylamine methyltransferase activity(GO:0043834) methanol-specific methylcobalamin:coenzyme M methyltransferase activity(GO:0043851) monomethylamine methyltransferase activity(GO:0043852) P-methyltransferase activity(GO:0051994) Se-methyltransferase activity(GO:0051995) 2-phytyl-1,4-naphthoquinone methyltransferase activity(GO:0052624) tRNA (uracil-2'-O-)-methyltransferase activity(GO:0052665) tRNA (cytosine-2'-O-)-methyltransferase activity(GO:0052666) phosphomethylethanolamine N-methyltransferase activity(GO:0052667) tRNA (cytosine-3-)-methyltransferase activity(GO:0052735) rRNA (cytosine-2'-O-)-methyltransferase activity(GO:0070677) rRNA (cytosine-N4-)-methyltransferase activity(GO:0071424) trihydroxyferuloyl spermidine O-methyltransferase activity(GO:0080012) |
| 0.0 | 0.7 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 1.1 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 1.4 | GO:0008565 | protein transporter activity(GO:0008565) |