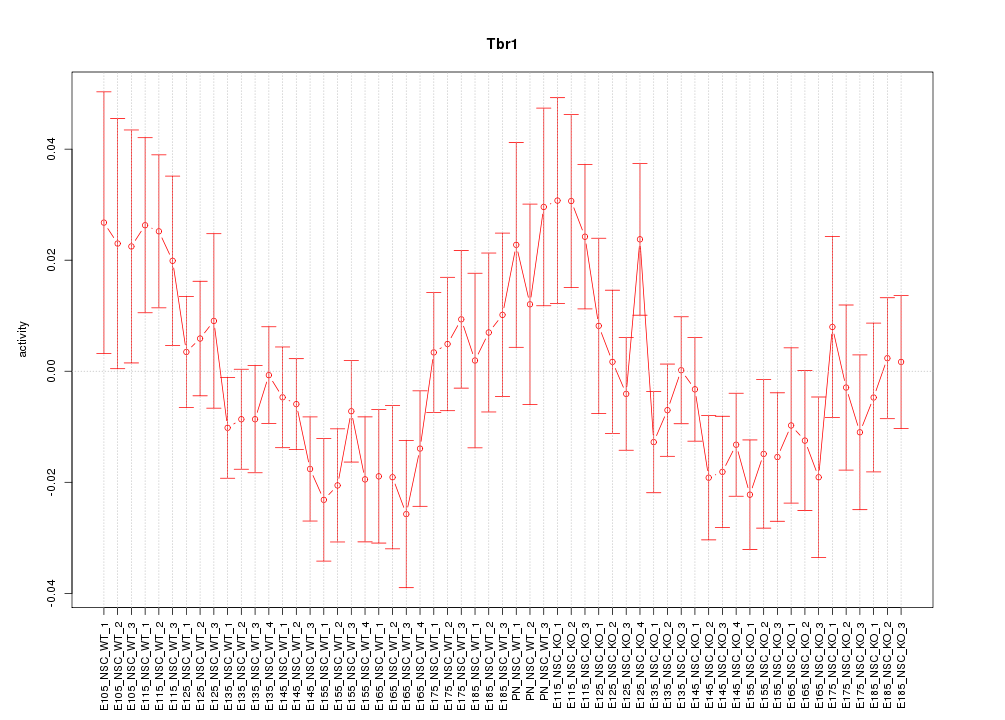
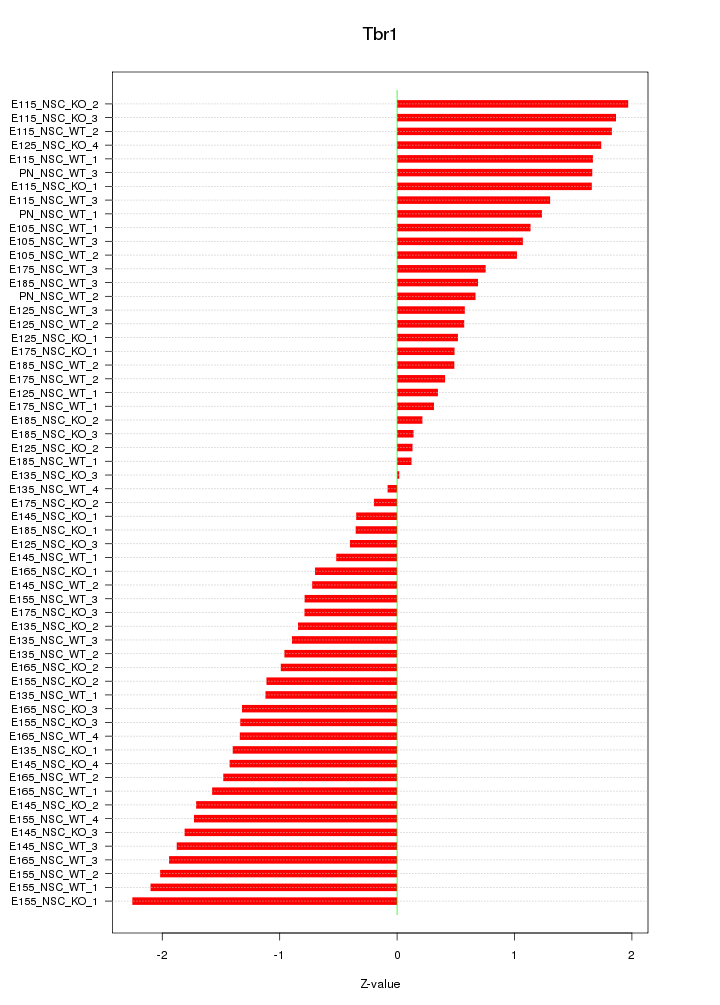
Motif ID: Tbr1
Z-value: 1.201

Transcription factors associated with Tbr1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Tbr1 | ENSMUSG00000035033.9 | Tbr1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tbr1 | mm10_v2_chr2_+_61804453_61804538 | -0.61 | 2.6e-07 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.6 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 2.2 | 8.9 | GO:0002339 | B cell selection(GO:0002339) |
| 2.1 | 6.4 | GO:1903232 | melanosome assembly(GO:1903232) |
| 2.1 | 6.3 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 1.9 | 7.8 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 1.9 | 9.4 | GO:0030091 | protein repair(GO:0030091) |
| 1.7 | 6.6 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 1.4 | 4.3 | GO:0030421 | defecation(GO:0030421) |
| 1.4 | 7.1 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 1.3 | 3.8 | GO:0045014 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 1.3 | 7.5 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 1.2 | 3.6 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 1.2 | 3.6 | GO:0019343 | cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) |
| 1.2 | 7.0 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 1.1 | 4.5 | GO:0032610 | interleukin-1 alpha production(GO:0032610) |
| 1.1 | 2.2 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) terminal web assembly(GO:1902896) |
| 1.1 | 7.7 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 1.1 | 3.3 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 1.0 | 3.1 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 1.0 | 3.0 | GO:0014028 | notochord formation(GO:0014028) |
| 0.9 | 4.7 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.9 | 2.8 | GO:0090526 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.9 | 3.8 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.9 | 2.8 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.9 | 3.5 | GO:0035934 | corticosterone secretion(GO:0035934) |
| 0.8 | 3.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.8 | 3.8 | GO:0019659 | lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.8 | 3.0 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.7 | 2.2 | GO:2001139 | negative regulation of postsynaptic membrane organization(GO:1901627) negative regulation of dendritic spine maintenance(GO:1902951) negative regulation of phospholipid efflux(GO:1902999) regulation of lipid transport across blood brain barrier(GO:1903000) negative regulation of lipid transport across blood brain barrier(GO:1903001) positive regulation of lipid transport across blood brain barrier(GO:1903002) negative regulation of phospholipid transport(GO:2001139) |
| 0.7 | 2.2 | GO:0030862 | neuroblast division in subventricular zone(GO:0021849) regulation of polarized epithelial cell differentiation(GO:0030860) positive regulation of polarized epithelial cell differentiation(GO:0030862) |
| 0.7 | 2.8 | GO:0035795 | negative regulation of mitochondrial membrane permeability(GO:0035795) |
| 0.7 | 3.4 | GO:0071038 | nuclear polyadenylation-dependent tRNA catabolic process(GO:0071038) |
| 0.7 | 4.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.6 | 2.6 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.6 | 1.9 | GO:0002586 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.6 | 2.6 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.6 | 1.9 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.6 | 2.5 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.6 | 1.8 | GO:1900673 | olefin metabolic process(GO:1900673) |
| 0.6 | 4.8 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.6 | 2.4 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.6 | 1.7 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.6 | 2.3 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.6 | 4.5 | GO:0051255 | spindle midzone assembly(GO:0051255) |
| 0.6 | 2.3 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.6 | 2.2 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.6 | 2.2 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.5 | 3.3 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.5 | 1.6 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) regulation of interleukin-6-mediated signaling pathway(GO:0070103) negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 0.5 | 3.8 | GO:0051461 | positive regulation of heat generation(GO:0031652) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.5 | 1.6 | GO:1990481 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) snRNA pseudouridine synthesis(GO:0031120) mRNA pseudouridine synthesis(GO:1990481) |
| 0.5 | 4.2 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.5 | 1.6 | GO:2000569 | T-helper 2 cell activation(GO:0035712) positive regulation of T-helper 17 type immune response(GO:2000318) regulation of T-helper 2 cell activation(GO:2000569) positive regulation of T-helper 2 cell activation(GO:2000570) |
| 0.5 | 1.6 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.5 | 2.6 | GO:0060161 | positive regulation of dopamine receptor signaling pathway(GO:0060161) |
| 0.5 | 2.0 | GO:1903998 | regulation of eating behavior(GO:1903998) |
| 0.5 | 1.5 | GO:0036233 | glycine import(GO:0036233) |
| 0.5 | 1.4 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.5 | 6.3 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.5 | 2.9 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.5 | 1.4 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) negative regulation of Fas signaling pathway(GO:1902045) |
| 0.5 | 0.9 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.5 | 3.3 | GO:2000781 | DNA dealkylation involved in DNA repair(GO:0006307) positive regulation of double-strand break repair(GO:2000781) |
| 0.5 | 2.3 | GO:0046645 | positive regulation of gamma-delta T cell differentiation(GO:0045588) positive regulation of gamma-delta T cell activation(GO:0046645) |
| 0.5 | 1.4 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.5 | 1.8 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.5 | 2.7 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.4 | 1.3 | GO:0002842 | T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) positive regulation of T cell mediated immune response to tumor cell(GO:0002842) |
| 0.4 | 1.3 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.4 | 1.8 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) resolution of recombination intermediates(GO:0071139) |
| 0.4 | 1.3 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.4 | 3.5 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.4 | 2.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.4 | 1.3 | GO:0021893 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 0.4 | 3.8 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.4 | 1.2 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.4 | 2.9 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.4 | 2.5 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.4 | 1.2 | GO:2000316 | negative regulation of interferon-gamma secretion(GO:1902714) regulation of T-helper 17 type immune response(GO:2000316) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.4 | 2.8 | GO:2001185 | regulation of CD8-positive, alpha-beta T cell activation(GO:2001185) |
| 0.4 | 1.6 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.4 | 1.2 | GO:0000050 | urea cycle(GO:0000050) regulation of amino acid import(GO:0010958) regulation of L-arginine import(GO:0010963) urea metabolic process(GO:0019627) |
| 0.4 | 0.4 | GO:2000811 | regulation of anoikis(GO:2000209) negative regulation of anoikis(GO:2000811) |
| 0.4 | 0.8 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.4 | 0.4 | GO:0003134 | BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) |
| 0.4 | 0.4 | GO:0032811 | negative regulation of epinephrine secretion(GO:0032811) |
| 0.4 | 1.2 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 0.4 | 2.2 | GO:0090394 | negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 0.4 | 4.6 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.4 | 7.8 | GO:0001913 | T cell mediated cytotoxicity(GO:0001913) |
| 0.3 | 8.0 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.3 | 1.4 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.3 | 2.4 | GO:0090399 | replicative senescence(GO:0090399) |
| 0.3 | 2.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.3 | 4.7 | GO:0060055 | angiogenesis involved in wound healing(GO:0060055) |
| 0.3 | 1.0 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.3 | 1.0 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.3 | 1.6 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.3 | 2.3 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.3 | 0.7 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.3 | 1.0 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.3 | 1.6 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.3 | 0.6 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.3 | 0.9 | GO:0007521 | muscle cell fate determination(GO:0007521) mammary placode formation(GO:0060596) |
| 0.3 | 0.9 | GO:2000256 | positive regulation of male germ cell proliferation(GO:2000256) |
| 0.3 | 2.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.3 | 0.9 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.3 | 1.5 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.3 | 1.2 | GO:0070814 | hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.3 | 2.7 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.3 | 1.2 | GO:0071726 | response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) |
| 0.3 | 0.9 | GO:0009812 | flavonoid metabolic process(GO:0009812) flavonoid biosynthetic process(GO:0009813) flavonoid glucuronidation(GO:0052696) |
| 0.3 | 2.9 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.3 | 3.2 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.3 | 0.9 | GO:0001803 | antibody-dependent cellular cytotoxicity(GO:0001788) type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 0.3 | 1.1 | GO:0006842 | tricarboxylic acid transport(GO:0006842) succinate transport(GO:0015744) citrate transport(GO:0015746) |
| 0.3 | 1.1 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.3 | 1.9 | GO:0009162 | nucleoside monophosphate catabolic process(GO:0009125) deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.3 | 1.6 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.3 | 3.5 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.3 | 1.0 | GO:0015871 | choline transport(GO:0015871) |
| 0.3 | 1.3 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.2 | 0.7 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 1.2 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.2 | 1.2 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.2 | 1.0 | GO:0036506 | maintenance of unfolded protein(GO:0036506) protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.2 | 2.6 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.2 | 2.1 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.2 | 3.0 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.2 | 1.6 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.2 | 2.0 | GO:0044130 | negative regulation of growth of symbiont in host(GO:0044130) negative regulation of growth of symbiont involved in interaction with host(GO:0044146) |
| 0.2 | 3.8 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.2 | 1.3 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.2 | 1.1 | GO:0010359 | regulation of anion channel activity(GO:0010359) |
| 0.2 | 0.6 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.2 | 1.9 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.2 | 1.5 | GO:0042693 | muscle cell fate commitment(GO:0042693) |
| 0.2 | 1.3 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.2 | 1.3 | GO:0098734 | protein depalmitoylation(GO:0002084) positive regulation of pinocytosis(GO:0048549) macromolecule depalmitoylation(GO:0098734) |
| 0.2 | 1.7 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.2 | 2.7 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.2 | 1.6 | GO:0033630 | positive regulation of cell adhesion mediated by integrin(GO:0033630) |
| 0.2 | 2.8 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.2 | 1.2 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.2 | 1.6 | GO:0097501 | stress response to metal ion(GO:0097501) |
| 0.2 | 5.6 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.2 | 0.2 | GO:0055001 | muscle cell development(GO:0055001) |
| 0.2 | 0.7 | GO:0018343 | protein farnesylation(GO:0018343) negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.2 | 1.3 | GO:0070458 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.2 | 0.6 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.2 | 2.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.2 | 1.7 | GO:0070633 | transepithelial transport(GO:0070633) |
| 0.2 | 1.6 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.2 | 0.7 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.2 | 2.0 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.2 | 0.3 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.2 | 1.4 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.2 | 0.5 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.2 | 2.6 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.2 | 1.0 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.2 | 1.3 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.2 | 0.7 | GO:0070305 | positive regulation of oocyte development(GO:0060282) response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) |
| 0.2 | 0.8 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.2 | 0.8 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.2 | 1.9 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.2 | 0.9 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.2 | 1.6 | GO:0030539 | male genitalia development(GO:0030539) |
| 0.2 | 1.4 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.2 | 0.8 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.2 | 2.4 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.2 | 0.6 | GO:0019287 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.2 | 0.5 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 0.6 | GO:0009181 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.1 | 1.2 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.1 | 4.0 | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway(GO:0040036) |
| 0.1 | 1.0 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.1 | 1.8 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.1 | 0.6 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 1.0 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 2.3 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.1 | 6.6 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 | 2.4 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.1 | 0.8 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.1 | 2.4 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 0.4 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.1 | 4.1 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.1 | 0.7 | GO:0042534 | tumor necrosis factor biosynthetic process(GO:0042533) regulation of tumor necrosis factor biosynthetic process(GO:0042534) |
| 0.1 | 0.9 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 0.4 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 0.4 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) T cell extravasation(GO:0072683) |
| 0.1 | 2.8 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.1 | 6.7 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.1 | 0.3 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.4 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.1 | 3.5 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.1 | 2.2 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 1.8 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.1 | 0.2 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.1 | 1.8 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 0.8 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 1.2 | GO:1901409 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 0.8 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 5.4 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.1 | 3.7 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 0.6 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 0.7 | GO:1903352 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.1 | 1.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 1.0 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 1.9 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 0.2 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 1.0 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 0.5 | GO:0071638 | cellular triglyceride homeostasis(GO:0035356) adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.1 | 0.6 | GO:1903887 | motile primary cilium assembly(GO:1903887) |
| 0.1 | 1.5 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.1 | 1.3 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.1 | 2.6 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 0.8 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.1 | 1.6 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.1 | 1.0 | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 0.5 | GO:0071404 | cellular response to lipoprotein particle stimulus(GO:0071402) cellular response to low-density lipoprotein particle stimulus(GO:0071404) positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 | 0.8 | GO:0032757 | positive regulation of interleukin-8 production(GO:0032757) |
| 0.1 | 1.3 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 1.7 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.1 | 3.7 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.1 | 0.7 | GO:1903894 | regulation of IRE1-mediated unfolded protein response(GO:1903894) |
| 0.1 | 0.3 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.1 | 0.8 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.1 | 0.7 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.1 | 1.9 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.1 | 0.2 | GO:0097274 | urea homeostasis(GO:0097274) |
| 0.1 | 3.4 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) |
| 0.1 | 0.2 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.1 | 3.0 | GO:0031050 | dsRNA fragmentation(GO:0031050) production of miRNAs involved in gene silencing by miRNA(GO:0035196) production of small RNA involved in gene silencing by RNA(GO:0070918) |
| 0.1 | 0.6 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.1 | 0.3 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 0.5 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.1 | 1.1 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.1 | 3.0 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.1 | 4.1 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.1 | 0.3 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.1 | 2.6 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 0.6 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 0.3 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.1 | 1.5 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.1 | 0.5 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 1.4 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.1 | 1.0 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 2.2 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.1 | 1.0 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.1 | 3.2 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 0.6 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.1 | 3.1 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.1 | 0.6 | GO:0046500 | S-adenosylmethionine metabolic process(GO:0046500) |
| 0.1 | 4.4 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.1 | 0.8 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.1 | 1.5 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.1 | 0.9 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 | 0.3 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 0.3 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.1 | 0.3 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.1 | 0.8 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 0.8 | GO:0052646 | alditol phosphate metabolic process(GO:0052646) |
| 0.1 | 0.9 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.2 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.0 | 3.5 | GO:0008584 | male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.0 | 0.2 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.1 | GO:0060693 | regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.0 | 1.3 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 1.4 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.2 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 1.9 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 | 0.5 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.8 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.6 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 1.1 | GO:0033014 | porphyrin-containing compound biosynthetic process(GO:0006779) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.0 | 0.4 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.4 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 1.4 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 2.1 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.3 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.4 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.1 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) ceramide transport(GO:0035627) |
| 0.0 | 4.8 | GO:0050658 | nucleic acid transport(GO:0050657) RNA transport(GO:0050658) |
| 0.0 | 0.4 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.2 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.7 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.3 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.6 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.3 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 1.3 | GO:2001022 | positive regulation of response to DNA damage stimulus(GO:2001022) |
| 0.0 | 2.5 | GO:1903749 | positive regulation of establishment of protein localization to mitochondrion(GO:1903749) positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.5 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.0 | 1.5 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.0 | 0.1 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.6 | GO:1901031 | regulation of response to reactive oxygen species(GO:1901031) |
| 0.0 | 0.1 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.0 | 0.2 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 1.9 | GO:0090305 | nucleic acid phosphodiester bond hydrolysis(GO:0090305) |
| 0.0 | 0.4 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.9 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.0 | GO:0001798 | type IIa hypersensitivity(GO:0001794) regulation of type IIa hypersensitivity(GO:0001796) positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type I hypersensitivity(GO:0001812) type II hypersensitivity(GO:0002445) positive regulation of inflammatory response to antigenic stimulus(GO:0002863) positive regulation of acute inflammatory response to antigenic stimulus(GO:0002866) positive regulation of hypersensitivity(GO:0002885) regulation of type II hypersensitivity(GO:0002892) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.0 | 0.1 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.3 | GO:1990090 | response to nerve growth factor(GO:1990089) cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.0 | 0.3 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.7 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.3 | GO:0000731 | DNA synthesis involved in DNA repair(GO:0000731) |
| 0.0 | 0.2 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 2.8 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.0 | 0.6 | GO:0001947 | heart looping(GO:0001947) |
| 0.0 | 0.4 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.0 | 0.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.7 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 1.0 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 0.3 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 2.2 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.2 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.3 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.0 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.4 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.0 | 2.7 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.0 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.1 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.0 | 0.1 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 1.1 | GO:0015698 | inorganic anion transport(GO:0015698) |
| 0.0 | 0.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.2 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.4 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.2 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.0 | 0.4 | GO:0045580 | regulation of T cell differentiation(GO:0045580) |
| 0.0 | 0.2 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.1 | GO:0002092 | positive regulation of receptor internalization(GO:0002092) |
| 0.0 | 0.4 | GO:0034101 | erythrocyte homeostasis(GO:0034101) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 10.7 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 2.4 | 7.2 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 1.6 | 4.8 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 1.3 | 8.9 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.9 | 4.4 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.8 | 4.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.8 | 2.4 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.7 | 2.2 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.7 | 4.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.7 | 7.5 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.6 | 1.9 | GO:0036125 | mitochondrial fatty acid beta-oxidation multienzyme complex(GO:0016507) fatty acid beta-oxidation multienzyme complex(GO:0036125) |
| 0.6 | 4.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.5 | 1.6 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.5 | 1.6 | GO:1990075 | stereocilia ankle link complex(GO:0002142) periciliary membrane compartment(GO:1990075) |
| 0.5 | 3.2 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.5 | 4.8 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.5 | 3.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.5 | 6.8 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.5 | 1.9 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.5 | 4.6 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.4 | 2.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.4 | 8.0 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.4 | 1.6 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.4 | 2.3 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.4 | 1.5 | GO:0005712 | chiasma(GO:0005712) |
| 0.4 | 1.8 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.3 | 2.4 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.3 | 3.7 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.3 | 3.9 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.3 | 1.9 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.3 | 2.5 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.3 | 1.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.3 | 7.0 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.3 | 3.1 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.3 | 2.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.2 | 0.7 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.2 | 0.7 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.2 | 1.0 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.2 | 3.4 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.2 | 0.9 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.2 | 9.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 1.3 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.2 | 1.9 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.2 | 0.8 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.2 | 0.4 | GO:0044299 | C-fiber(GO:0044299) |
| 0.2 | 1.5 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.2 | 1.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.2 | 1.7 | GO:0034709 | methylosome(GO:0034709) |
| 0.2 | 2.1 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.2 | 0.5 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.2 | 0.5 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.2 | 0.9 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.2 | 1.4 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.1 | 3.6 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 0.6 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 1.0 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 0.6 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.4 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.1 | 7.8 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 1.9 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 1.9 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 1.8 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 1.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.4 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 1.0 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 7.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 1.6 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 1.4 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 1.9 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.1 | 1.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 1.0 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 1.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.1 | 4.8 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 1.8 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 0.3 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 1.0 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 1.3 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.1 | 1.3 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 0.9 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 1.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 1.8 | GO:0030894 | replisome(GO:0030894) |
| 0.1 | 0.9 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 2.2 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 5.3 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 3.7 | GO:0043235 | receptor complex(GO:0043235) |
| 0.1 | 0.7 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 1.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.3 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.1 | 3.6 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 2.0 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.5 | GO:0030677 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.1 | 1.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 1.7 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 12.7 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 6.3 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 1.8 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 0.9 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 0.8 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 1.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.6 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 0.2 | GO:0070449 | elongin complex(GO:0070449) |
| 0.1 | 0.8 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 2.7 | GO:0000313 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.1 | 6.5 | GO:0015934 | large ribosomal subunit(GO:0015934) |
| 0.1 | 0.6 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 0.5 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 3.2 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.1 | 0.3 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 3.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 0.6 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.5 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.3 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.8 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 1.7 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.6 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 1.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 8.6 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.6 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 1.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 14.7 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 5.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.2 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.5 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 1.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.5 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 1.4 | GO:0044391 | small ribosomal subunit(GO:0015935) cytosolic small ribosomal subunit(GO:0022627) ribosomal subunit(GO:0044391) |
| 0.0 | 1.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 3.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.0 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.5 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 8.8 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 5.5 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 1.0 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.6 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 1.3 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 1.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.4 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.2 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 2.8 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 1.1 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 1.2 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 4.1 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 2.9 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.0 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 0.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.6 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 2.2 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.0 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.8 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.2 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 10.7 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 2.1 | 6.4 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 1.7 | 10.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 1.7 | 11.7 | GO:0070191 | methionine-R-sulfoxide reductase activity(GO:0070191) |
| 1.6 | 8.0 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 1.4 | 7.0 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 1.4 | 4.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 1.3 | 6.7 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 1.3 | 4.0 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 1.2 | 10.8 | GO:0015643 | toxic substance binding(GO:0015643) |
| 1.1 | 8.4 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 1.0 | 3.1 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 1.0 | 3.0 | GO:0005118 | sevenless binding(GO:0005118) |
| 1.0 | 3.8 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.9 | 7.3 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.9 | 2.7 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.9 | 7.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.8 | 4.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.8 | 3.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.7 | 2.2 | GO:0046911 | hydroxyapatite binding(GO:0046848) metal chelating activity(GO:0046911) phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.7 | 2.1 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.7 | 2.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.7 | 2.7 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.6 | 2.6 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.6 | 7.5 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.6 | 5.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.6 | 4.7 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.6 | 2.8 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.5 | 1.6 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) oncostatin-M receptor activity(GO:0004924) |
| 0.5 | 2.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.5 | 3.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.5 | 4.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.5 | 2.6 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.5 | 6.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.5 | 1.5 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.5 | 1.5 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.5 | 1.9 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.5 | 2.8 | GO:0070728 | leucine binding(GO:0070728) |
| 0.5 | 2.3 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.5 | 1.4 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.5 | 15.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.4 | 1.8 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.4 | 1.3 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.4 | 2.6 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.4 | 1.3 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.4 | 1.7 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.4 | 1.2 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) |
| 0.4 | 1.6 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.4 | 1.6 | GO:0086038 | calcium:sodium antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086038) |
| 0.4 | 1.6 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.4 | 1.2 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.4 | 1.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.4 | 1.1 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.4 | 1.5 | GO:0038049 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.4 | 3.6 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.3 | 7.7 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.3 | 3.5 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.3 | 2.3 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.3 | 1.6 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.3 | 1.9 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.3 | 4.8 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.3 | 0.9 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.3 | 0.9 | GO:0019863 | IgE binding(GO:0019863) |
| 0.3 | 1.2 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.3 | 0.9 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.3 | 1.2 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.3 | 0.9 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) IgG receptor activity(GO:0019770) |
| 0.3 | 0.8 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.3 | 2.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.3 | 0.8 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.3 | 1.6 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.3 | 1.0 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.3 | 1.3 | GO:0043842 | Kdo transferase activity(GO:0043842) |
| 0.3 | 7.1 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.2 | 0.7 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.2 | 2.5 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.2 | 4.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 1.9 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.2 | 2.1 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.2 | 0.9 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.2 | 0.9 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.2 | 1.4 | GO:0003896 | DNA primase activity(GO:0003896) CTP:2,3-di-O-geranylgeranyl-sn-glycero-1-phosphate cytidyltransferase activity(GO:0043338) phospholactate guanylyltransferase activity(GO:0043814) ATP:coenzyme F420 adenylyltransferase activity(GO:0043910) UDP-N-acetylgalactosamine diphosphorylase activity(GO:0052630) |
| 0.2 | 10.5 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.2 | 4.8 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.2 | 1.3 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.2 | 1.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.2 | 10.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.2 | 0.6 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.2 | 4.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 1.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.2 | 2.0 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.2 | 3.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 0.8 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.2 | 0.8 | GO:0042806 | fucose binding(GO:0042806) oligosaccharide binding(GO:0070492) |
| 0.2 | 1.0 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.2 | 1.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.2 | 1.0 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.2 | 2.7 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.2 | 0.6 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.2 | 0.9 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.2 | 0.6 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.2 | 2.6 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.2 | 1.6 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.2 | 1.1 | GO:0034916 | 4-methyloctanoyl-CoA dehydrogenase activity(GO:0034580) naphthyl-2-methyl-succinyl-CoA dehydrogenase activity(GO:0034845) 2-methylhexanoyl-CoA dehydrogenase activity(GO:0034916) propionyl-CoA dehydrogenase activity(GO:0043820) thiol-driven fumarate reductase activity(GO:0043830) coenzyme F420-dependent 2,4,6-trinitrophenol reductase activity(GO:0052758) coenzyme F420-dependent 2,4,6-trinitrophenol hydride reductase activity(GO:0052759) coenzyme F420-dependent 2,4-dinitrophenol reductase activity(GO:0052760) |
| 0.2 | 1.3 | GO:0043559 | insulin binding(GO:0043559) |
| 0.2 | 3.8 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.2 | 2.2 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.2 | 1.9 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.2 | 3.0 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.2 | 3.5 | GO:0044653 | trehalase activity(GO:0015927) dextrin alpha-glucosidase activity(GO:0044653) starch alpha-glucosidase activity(GO:0044654) beta-glucanase activity(GO:0052736) beta-6-sulfate-N-acetylglucosaminidase activity(GO:0052769) glucan endo-1,4-beta-glucosidase activity(GO:0052859) |
| 0.1 | 1.2 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) DNA polymerase activity(GO:0034061) |
| 0.1 | 4.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.3 | GO:0051393 | retinoic acid binding(GO:0001972) alpha-actinin binding(GO:0051393) |
| 0.1 | 1.3 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 1.7 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 1.0 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 2.7 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 0.4 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 1.0 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 2.5 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 6.9 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 1.3 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.6 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 1.2 | GO:0004889 | acetylcholine-activated cation-selective channel activity(GO:0004889) acetylcholine binding(GO:0042166) |
| 0.1 | 1.2 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 5.3 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 1.9 | GO:0031702 | angiotensin receptor binding(GO:0031701) type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 1.3 | GO:0042556 | cobinamide kinase activity(GO:0008819) phytol kinase activity(GO:0010276) phenol kinase activity(GO:0018720) cyclin-dependent protein kinase activating kinase regulator activity(GO:0019914) inositol tetrakisphosphate 2-kinase activity(GO:0032942) heptose 7-phosphate kinase activity(GO:0033785) aminoglycoside phosphotransferase activity(GO:0034071) eukaryotic elongation factor-2 kinase regulator activity(GO:0042556) eukaryotic elongation factor-2 kinase activator activity(GO:0042557) LPPG:FO 2-phospho-L-lactate transferase activity(GO:0043743) cytidine kinase activity(GO:0043771) glycerate 2-kinase activity(GO:0043798) (S)-lactate 2-kinase activity(GO:0043841) phosphoserine:homoserine phosphotransferase activity(GO:0043899) L-seryl-tRNA(Sec) kinase activity(GO:0043915) phosphocholine transferase activity(GO:0044605) GTP-dependent polynucleotide kinase activity(GO:0051735) farnesol kinase activity(GO:0052668) CTP:2-trans,-6-trans-farnesol kinase activity(GO:0052669) geraniol kinase activity(GO:0052670) geranylgeraniol kinase activity(GO:0052671) CTP:geranylgeraniol kinase activity(GO:0052672) prenol kinase activity(GO:0052673) 1-phosphatidylinositol-5-kinase activity(GO:0052810) 1-phosphatidylinositol-3-phosphate 4-kinase activity(GO:0052811) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.1 | 0.8 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 2.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.8 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.6 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.1 | 0.8 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.9 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.1 | 1.9 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 2.3 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.3 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 3.1 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) exoribonuclease activity, producing 5'-phosphomonoesters(GO:0016896) |
| 0.1 | 0.7 | GO:0015189 | L-ornithine transmembrane transporter activity(GO:0000064) arginine transmembrane transporter activity(GO:0015181) L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 1.1 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.1 | 3.8 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 1.0 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.1 | 0.3 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.1 | 0.8 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 6.4 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 1.6 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 1.8 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 1.7 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 1.0 | GO:0070636 | single-strand selective uracil DNA N-glycosylase activity(GO:0017065) nicotinamide riboside hydrolase activity(GO:0070635) nicotinic acid riboside hydrolase activity(GO:0070636) deoxyribonucleoside 5'-monophosphate N-glycosidase activity(GO:0070694) |
| 0.1 | 0.2 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.1 | 1.8 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 1.6 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 1.2 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.1 | 1.4 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 0.3 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 0.2 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 1.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 6.8 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 0.3 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.1 | 1.6 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 0.9 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 1.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 1.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 3.1 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 1.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 0.7 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 1.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 1.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.8 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.9 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 1.5 | GO:0045502 | dynein binding(GO:0045502) |
| 0.1 | 4.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.7 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.2 | GO:0001094 | RNA polymerase II basal transcription factor binding(GO:0001091) TFIID-class transcription factor binding(GO:0001094) |
| 0.1 | 0.2 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.1 | 1.7 | GO:0001067 | regulatory region DNA binding(GO:0000975) regulatory region nucleic acid binding(GO:0001067) |
| 0.1 | 0.3 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.1 | 1.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 3.4 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.1 | 0.4 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.1 | 0.9 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 1.4 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 0.4 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 1.4 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.1 | 0.5 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 1.3 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.2 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.0 | 1.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.5 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 4.1 | GO:0008234 | cysteine-type peptidase activity(GO:0008234) |
| 0.0 | 0.5 | GO:0008758 | thiamine-pyrophosphatase activity(GO:0004787) UDP-2,3-diacylglucosamine hydrolase activity(GO:0008758) dATP pyrophosphohydrolase activity(GO:0008828) dihydroneopterin monophosphate phosphatase activity(GO:0019176) dihydroneopterin triphosphate pyrophosphohydrolase activity(GO:0019177) dTTP diphosphatase activity(GO:0036218) phosphocholine hydrolase activity(GO:0044606) |
| 0.0 | 0.2 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 3.1 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 1.5 | GO:0008748 | N-ethylmaleimide reductase activity(GO:0008748) reduced coenzyme F420 dehydrogenase activity(GO:0043738) sulfur oxygenase reductase activity(GO:0043826) malolactic enzyme activity(GO:0043883) epoxyqueuosine reductase activity(GO:0052693) |
| 0.0 | 0.7 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.9 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 3.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.7 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.1 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 2.3 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.4 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.2 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.8 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.6 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.4 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 1.2 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.4 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 1.2 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 4.3 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 2.0 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 1.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.8 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 1.9 | GO:0043851 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) rRNA (uridine-2'-O-)-methyltransferase activity(GO:0008650) rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) selenocysteine methyltransferase activity(GO:0016205) rRNA (adenine) methyltransferase activity(GO:0016433) rRNA (cytosine) methyltransferase activity(GO:0016434) rRNA (guanine) methyltransferase activity(GO:0016435) 1-phenanthrol methyltransferase activity(GO:0018707) protein-arginine N5-methyltransferase activity(GO:0019702) dimethylarsinite methyltransferase activity(GO:0034541) 4,5-dihydroxybenzo(a)pyrene methyltransferase activity(GO:0034807) 1-hydroxypyrene methyltransferase activity(GO:0034931) 1-hydroxy-6-methoxypyrene methyltransferase activity(GO:0034933) demethylmenaquinone methyltransferase activity(GO:0043770) cobalt-precorrin-6B C5-methyltransferase activity(GO:0043776) cobalt-precorrin-7 C15-methyltransferase activity(GO:0043777) cobalt-precorrin-5B C1-methyltransferase activity(GO:0043780) cobalt-precorrin-3 C17-methyltransferase activity(GO:0043782) dimethylamine methyltransferase activity(GO:0043791) hydroxyneurosporene-O-methyltransferase activity(GO:0043803) tRNA (adenine-57, 58-N(1)-) methyltransferase activity(GO:0043827) methylamine-specific methylcobalamin:coenzyme M methyltransferase activity(GO:0043833) trimethylamine methyltransferase activity(GO:0043834) methanol-specific methylcobalamin:coenzyme M methyltransferase activity(GO:0043851) monomethylamine methyltransferase activity(GO:0043852) P-methyltransferase activity(GO:0051994) Se-methyltransferase activity(GO:0051995) 2-phytyl-1,4-naphthoquinone methyltransferase activity(GO:0052624) tRNA (uracil-2'-O-)-methyltransferase activity(GO:0052665) tRNA (cytosine-2'-O-)-methyltransferase activity(GO:0052666) phosphomethylethanolamine N-methyltransferase activity(GO:0052667) tRNA (cytosine-3-)-methyltransferase activity(GO:0052735) rRNA (cytosine-2'-O-)-methyltransferase activity(GO:0070677) rRNA (cytosine-N4-)-methyltransferase activity(GO:0071424) trihydroxyferuloyl spermidine O-methyltransferase activity(GO:0080012) |
| 0.0 | 0.1 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.3 | GO:0005253 | anion channel activity(GO:0005253) |
| 0.0 | 1.2 | GO:0015108 | chloride transmembrane transporter activity(GO:0015108) |
| 0.0 | 0.7 | GO:0005496 | steroid binding(GO:0005496) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.6 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.0 | 0.7 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.6 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 6.5 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.0 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) |
| 0.0 | 0.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.0 | 0.3 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 1.2 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.3 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.2 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.0 | 0.5 | GO:0000049 | tRNA binding(GO:0000049) |