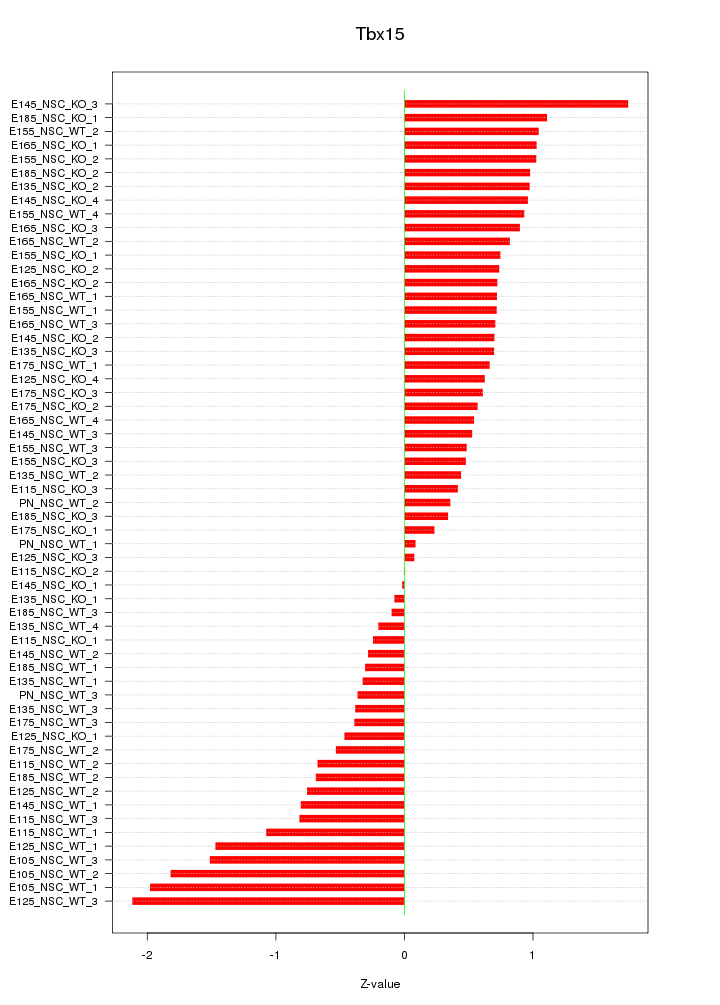
Motif ID: Tbx15
Z-value: 0.838
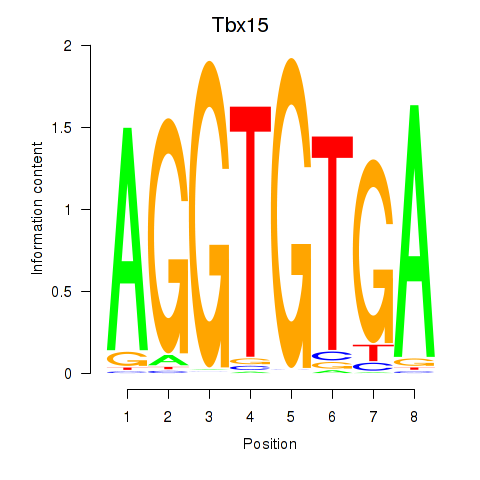
Transcription factors associated with Tbx15:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Tbx15 | ENSMUSG00000027868.5 | Tbx15 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 15.2 | GO:0030827 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 4.0 | 12.0 | GO:0038095 | positive regulation of mast cell cytokine production(GO:0032765) Fc-epsilon receptor signaling pathway(GO:0038095) |
| 2.8 | 8.4 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 2.4 | 7.2 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 2.3 | 4.7 | GO:0032763 | regulation of mast cell cytokine production(GO:0032763) |
| 1.9 | 7.7 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 1.8 | 5.3 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 1.6 | 4.8 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 1.6 | 6.2 | GO:0099624 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) atrial cardiac muscle cell membrane repolarization(GO:0099624) |
| 1.5 | 4.4 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 1.4 | 4.3 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 1.3 | 3.9 | GO:1902277 | negative regulation of pancreatic amylase secretion(GO:1902277) |
| 1.3 | 6.3 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 1.2 | 5.9 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 1.2 | 3.5 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 1.1 | 1.1 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 1.0 | 4.8 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.9 | 2.8 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.9 | 4.4 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.7 | 2.8 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.7 | 2.8 | GO:0060745 | mammary gland branching involved in pregnancy(GO:0060745) |
| 0.7 | 2.0 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.7 | 4.7 | GO:0034047 | regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.6 | 3.9 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.6 | 3.7 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.6 | 2.4 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.6 | 1.7 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) ceramide transport(GO:0035627) |
| 0.5 | 2.7 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.5 | 5.3 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.5 | 2.6 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.5 | 3.6 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.5 | 1.5 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.5 | 6.0 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.5 | 1.8 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.4 | 3.0 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.4 | 7.6 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.4 | 7.4 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.4 | 1.6 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 0.4 | 2.3 | GO:1903056 | regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.4 | 3.3 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.4 | 2.5 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.3 | 3.7 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.3 | 3.4 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.3 | 1.2 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.3 | 3.3 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.3 | 0.9 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.3 | 5.3 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.3 | 1.5 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.3 | 1.6 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.3 | 1.6 | GO:1903847 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.3 | 1.0 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.2 | 1.2 | GO:0044838 | cell quiescence(GO:0044838) |
| 0.2 | 1.4 | GO:0036006 | response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.2 | 4.2 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.2 | 1.6 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.2 | 5.1 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 1.8 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.2 | 4.1 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.2 | 0.8 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.2 | 1.9 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.2 | 3.4 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.2 | 0.4 | GO:0045963 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.2 | 2.7 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.2 | 1.1 | GO:1901524 | regulation of macromitophagy(GO:1901524) negative regulation of macromitophagy(GO:1901525) |
| 0.2 | 2.0 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.2 | 2.0 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.2 | 1.2 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.2 | 1.4 | GO:0036506 | maintenance of unfolded protein(GO:0036506) protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.2 | 0.7 | GO:0015871 | choline transport(GO:0015871) |
| 0.2 | 3.8 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.2 | 1.1 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.2 | 1.2 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.2 | 0.5 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.2 | 5.1 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 2.4 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.1 | 4.0 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 0.4 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.6 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.1 | 2.9 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 1.4 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.1 | 1.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.6 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.1 | 2.2 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.1 | 5.3 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.1 | 0.5 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.4 | GO:2000501 | natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.1 | 1.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 1.3 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 1.4 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.1 | 2.9 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.1 | 9.1 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.1 | 6.0 | GO:0035904 | aorta development(GO:0035904) |
| 0.1 | 0.4 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.1 | 0.5 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.1 | 3.7 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.1 | 1.5 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 0.6 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.7 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 4.5 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 3.0 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.1 | 0.4 | GO:0032275 | luteinizing hormone secretion(GO:0032275) positive regulation of gonadotropin secretion(GO:0032278) |
| 0.1 | 1.2 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 0.3 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.1 | 0.5 | GO:1903861 | regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 2.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 1.4 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 2.0 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 1.4 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.1 | 0.4 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.1 | 0.6 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.1 | 1.6 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.1 | 0.3 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.1 | 1.4 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 0.2 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.1 | 0.2 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.1 | 0.3 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 | 0.4 | GO:1903352 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.1 | 1.3 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.1 | 0.5 | GO:2001197 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.1 | 0.8 | GO:0030813 | positive regulation of nucleotide catabolic process(GO:0030813) positive regulation of glycolytic process(GO:0045821) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) |
| 0.1 | 0.8 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.1 | 2.3 | GO:1901381 | positive regulation of potassium ion transmembrane transport(GO:1901381) |
| 0.1 | 0.2 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.1 | 1.0 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 | 1.4 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.1 | 8.8 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.1 | 1.0 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.1 | 0.7 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.3 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 3.7 | GO:0030641 | regulation of cellular pH(GO:0030641) |
| 0.0 | 1.7 | GO:0015844 | monoamine transport(GO:0015844) |
| 0.0 | 0.5 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 0.6 | GO:0050995 | negative regulation of lipid catabolic process(GO:0050995) |
| 0.0 | 0.8 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 2.0 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 1.7 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.5 | GO:0042182 | ketone catabolic process(GO:0042182) |
| 0.0 | 0.4 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.0 | 0.7 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 | 1.2 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.0 | 2.1 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 1.2 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.0 | 5.0 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.1 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.0 | 0.8 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.2 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.0 | 0.4 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.1 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.2 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.0 | 1.0 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 2.3 | GO:0015758 | glucose transport(GO:0015758) |
| 0.0 | 0.7 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.3 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.0 | 0.4 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 2.1 | GO:0001824 | blastocyst development(GO:0001824) |
| 0.0 | 3.1 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.0 | 0.4 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.2 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.0 | 0.4 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.2 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.5 | GO:0042130 | negative regulation of T cell proliferation(GO:0042130) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 15.2 | GO:0044327 | dendritic spine head(GO:0044327) |
| 1.4 | 7.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 1.2 | 3.7 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.8 | 7.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.7 | 4.0 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.6 | 5.7 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.5 | 11.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.4 | 4.5 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.4 | 0.4 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.4 | 1.3 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.4 | 2.8 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.3 | 2.0 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.3 | 10.9 | GO:0030673 | axolemma(GO:0030673) |
| 0.3 | 3.4 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.3 | 1.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.3 | 3.4 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.3 | 2.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.3 | 15.3 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.3 | 5.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.3 | 1.7 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.2 | 4.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 3.6 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.2 | 2.7 | GO:0043196 | varicosity(GO:0043196) |
| 0.2 | 1.4 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.2 | 3.7 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 2.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 5.9 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 2.0 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 0.5 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 2.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 5.0 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 3.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 1.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 6.8 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 6.8 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 1.1 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 2.3 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 0.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 4.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 0.7 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 1.8 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 5.6 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 0.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 5.8 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 1.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.5 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 0.8 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 8.4 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.4 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 1.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.5 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 2.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 3.1 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 1.5 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 2.4 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 5.9 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 8.6 | GO:0099503 | secretory vesicle(GO:0099503) |
| 0.0 | 0.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.5 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 1.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.4 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.9 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 2.8 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 0.3 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 1.7 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.2 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 1.5 | 4.4 | GO:0031752 | D3 dopamine receptor binding(GO:0031750) D5 dopamine receptor binding(GO:0031752) |
| 1.4 | 5.6 | GO:0086038 | calcium:sodium antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086038) |
| 1.4 | 9.9 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 1.3 | 10.1 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 1.2 | 3.7 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 1.2 | 3.6 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 1.2 | 4.8 | GO:0004096 | catalase activity(GO:0004096) |
| 0.9 | 13.8 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.8 | 7.6 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.8 | 4.2 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.8 | 2.4 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.8 | 3.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.7 | 3.7 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.7 | 2.7 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.6 | 12.3 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.6 | 1.7 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.5 | 6.5 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.5 | 4.9 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.5 | 1.6 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.5 | 3.0 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.5 | 1.5 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.4 | 3.3 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.4 | 1.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.3 | 4.7 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.2 | 3.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.2 | 3.8 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.2 | 3.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 4.3 | GO:0050253 | prenylcysteine methylesterase activity(GO:0010296) 1-oxa-2-oxocycloheptane lactonase activity(GO:0018731) sulfolactone hydrolase activity(GO:0018732) butyrolactone hydrolase activity(GO:0018734) endosulfan lactone lactonase activity(GO:0034892) L-ascorbate 6-phosphate lactonase activity(GO:0035460) Ser-tRNA(Thr) hydrolase activity(GO:0043905) Ala-tRNA(Pro) hydrolase activity(GO:0043906) Cys-tRNA(Pro) hydrolase activity(GO:0043907) Ser(Gly)-tRNA(Ala) hydrolase activity(GO:0043908) all-trans-retinyl-palmitate hydrolase, all-trans-retinol forming activity(GO:0047376) retinyl-palmitate esterase activity(GO:0050253) mannosyl-oligosaccharide 1,6-alpha-mannosidase activity(GO:0052767) mannosyl-oligosaccharide 1,3-alpha-mannosidase activity(GO:0052768) methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.2 | 0.9 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.2 | 7.7 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.2 | 1.5 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.2 | 1.4 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.2 | 3.2 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.2 | 1.4 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.2 | 2.7 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.2 | 1.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.2 | 0.2 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.2 | 2.1 | GO:0044213 | intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.2 | 0.5 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.2 | 0.5 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.2 | 0.7 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.2 | 0.7 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.2 | 0.8 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 1.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 0.8 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 2.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 5.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 6.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.4 | GO:0036361 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.1 | 1.4 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 0.9 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 0.4 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.1 | 2.4 | GO:0043747 | protein-N-terminal asparagine amidohydrolase activity(GO:0008418) UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase activity(GO:0008759) iprodione amidohydrolase activity(GO:0018748) (3,5-dichlorophenylurea)acetate amidohydrolase activity(GO:0018749) 4'-(2-hydroxyisopropyl)phenylurea amidohydrolase activity(GO:0034571) didemethylisoproturon amidohydrolase activity(GO:0034573) N-isopropylacetanilide amidohydrolase activity(GO:0034576) N-cyclohexylformamide amidohydrolase activity(GO:0034781) isonicotinic acid hydrazide hydrolase activity(GO:0034876) cis-aconitamide amidase activity(GO:0034882) gamma-N-formylaminovinylacetate hydrolase activity(GO:0034885) N2-acetyl-L-lysine deacetylase activity(GO:0043747) O-succinylbenzoate synthase activity(GO:0043748) indoleacetamide hydrolase activity(GO:0043864) N-acetylcitrulline deacetylase activity(GO:0043909) N-acetylgalactosamine-6-phosphate deacetylase activity(GO:0047419) diacetylchitobiose deacetylase activity(GO:0052773) chitooligosaccharide deacetylase activity(GO:0052790) |
| 0.1 | 5.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 1.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.1 | 2.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 0.6 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.1 | 0.3 | GO:0015217 | ADP transmembrane transporter activity(GO:0015217) coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.1 | 1.2 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 2.0 | GO:0031690 | adrenergic receptor binding(GO:0031690) |
| 0.1 | 2.2 | GO:0070628 | proteasome binding(GO:0070628) ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 0.3 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.1 | 3.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 2.9 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 0.3 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.1 | 2.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 6.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 6.0 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 0.3 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.1 | 3.4 | GO:0008748 | N-ethylmaleimide reductase activity(GO:0008748) reduced coenzyme F420 dehydrogenase activity(GO:0043738) sulfur oxygenase reductase activity(GO:0043826) malolactic enzyme activity(GO:0043883) epoxyqueuosine reductase activity(GO:0052693) |
| 0.1 | 1.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.4 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 2.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.2 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) |
| 0.1 | 1.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 4.8 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 2.0 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.4 | GO:0015189 | L-ornithine transmembrane transporter activity(GO:0000064) arginine transmembrane transporter activity(GO:0015181) L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 7.9 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 1.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 2.0 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 2.2 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 1.3 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.1 | 0.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 1.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 11.4 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.5 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.4 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.5 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 1.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.0 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.9 | GO:0008308 | voltage-gated anion channel activity(GO:0008308) |
| 0.0 | 1.1 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 2.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.8 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.2 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.0 | 0.7 | GO:0035380 | C-3 sterol dehydrogenase (C-4 sterol decarboxylase) activity(GO:0000252) mevaldate reductase activity(GO:0004495) gluconate dehydrogenase activity(GO:0008875) epoxide dehydrogenase activity(GO:0018451) 5-exo-hydroxycamphor dehydrogenase activity(GO:0018452) 2-hydroxytetrahydrofuran dehydrogenase activity(GO:0018453) acetoin dehydrogenase activity(GO:0019152) phenylcoumaran benzylic ether reductase activity(GO:0032442) D-xylose:NADP reductase activity(GO:0032866) L-arabinose:NADP reductase activity(GO:0032867) D-arabinitol dehydrogenase, D-ribulose forming (NADP+) activity(GO:0033709) (R)-(-)-1,2,3,4-tetrahydronaphthol dehydrogenase activity(GO:0034831) 3-hydroxymenthone dehydrogenase activity(GO:0034840) very long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0035380) dihydrotestosterone 17-beta-dehydrogenase activity(GO:0035410) (R)-2-hydroxyisocaproate dehydrogenase activity(GO:0043713) L-arabinose 1-dehydrogenase (NADP+) activity(GO:0044103) L-xylulose reductase (NAD+) activity(GO:0044105) 3-ketoglucose-reductase activity(GO:0048258) D-arabinitol dehydrogenase, D-xylulose forming (NADP+) activity(GO:0052677) |
| 0.0 | 0.2 | GO:0043842 | Kdo transferase activity(GO:0043842) |
| 0.0 | 7.8 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.5 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 1.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 1.2 | GO:0004120 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) photoreceptor cyclic-nucleotide phosphodiesterase activity(GO:0004120) 7,8-dihydro-D-neopterin 2',3'-cyclic phosphate phosphodiesterase activity(GO:0044688) inositol phosphosphingolipid phospholipase activity(GO:0052712) inositol phosphorylceramide phospholipase activity(GO:0052713) mannosyl-inositol phosphorylceramide phospholipase activity(GO:0052714) mannosyl-diinositol phosphorylceramide phospholipase activity(GO:0052715) |
| 0.0 | 0.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 1.3 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.1 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.5 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.3 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.8 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.6 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 1.3 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 1.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.8 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.0 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 1.4 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 2.9 | GO:0000982 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) |
| 0.0 | 2.5 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.8 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 6.2 | GO:0019900 | kinase binding(GO:0019900) |
| 0.0 | 1.1 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |