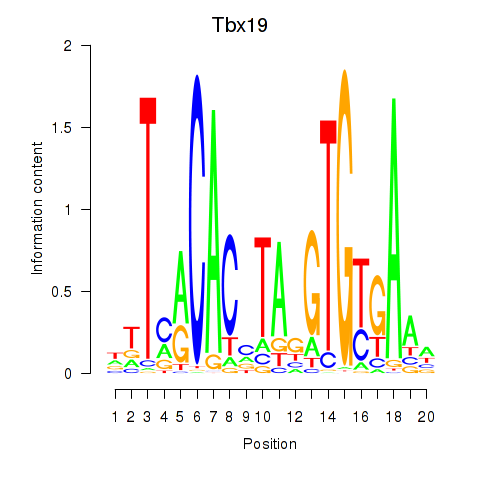
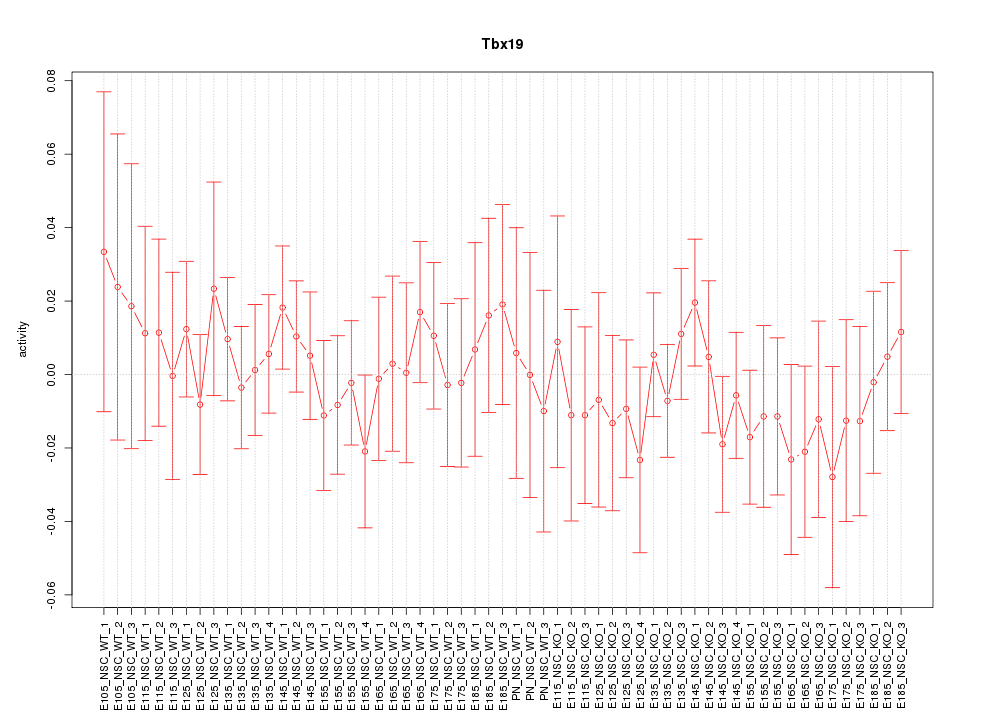
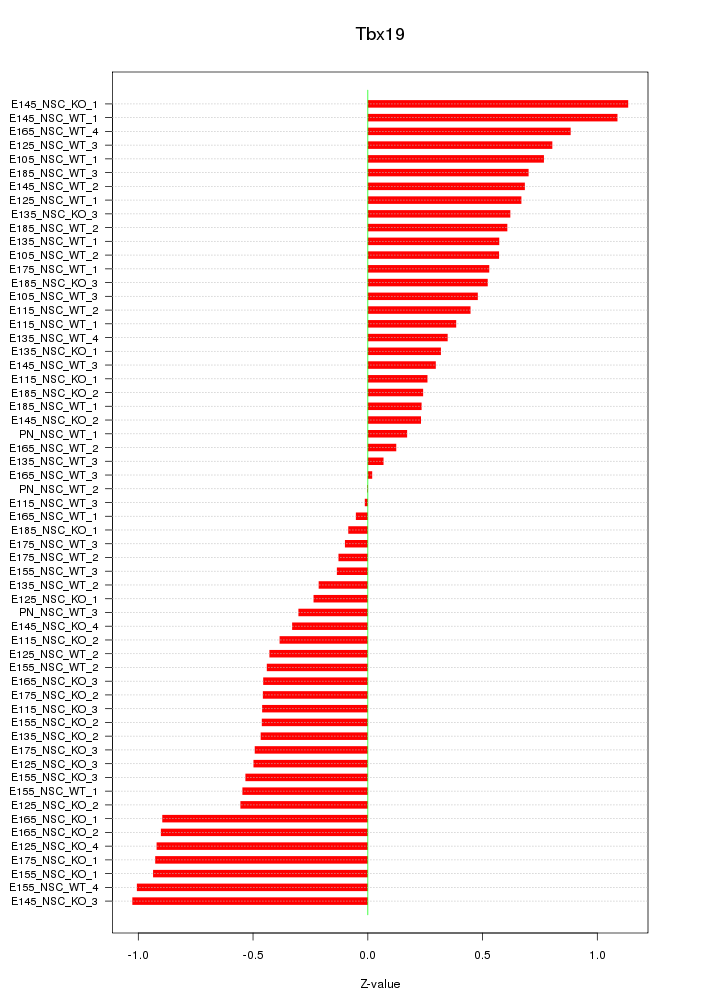
| 0.6 |
3.1 |
GO:0002457 |
T cell antigen processing and presentation(GO:0002457) |
| 0.5 |
2.0 |
GO:1902990 |
mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.4 |
1.1 |
GO:0010958 |
regulation of amino acid import(GO:0010958) regulation of L-arginine import(GO:0010963) |
| 0.3 |
1.2 |
GO:0071033 |
nuclear retention of pre-mRNA at the site of transcription(GO:0071033) CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.3 |
0.9 |
GO:0006420 |
arginyl-tRNA aminoacylation(GO:0006420) |
| 0.2 |
0.7 |
GO:0006578 |
amino-acid betaine biosynthetic process(GO:0006578) |
| 0.2 |
0.7 |
GO:0002538 |
arachidonic acid metabolite production involved in inflammatory response(GO:0002538) olefin metabolic process(GO:1900673) |
| 0.2 |
0.7 |
GO:0097278 |
complement-dependent cytotoxicity(GO:0097278) |
| 0.2 |
0.7 |
GO:0031590 |
wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.2 |
0.9 |
GO:0048304 |
positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.2 |
1.3 |
GO:1903690 |
negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.2 |
0.6 |
GO:0045819 |
positive regulation of glycogen catabolic process(GO:0045819) |
| 0.2 |
1.2 |
GO:0048934 |
peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.2 |
1.1 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.2 |
1.7 |
GO:0045719 |
negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.2 |
0.8 |
GO:0048631 |
regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.1 |
0.7 |
GO:0035372 |
protein localization to microtubule(GO:0035372) |
| 0.1 |
0.5 |
GO:0097527 |
necroptotic signaling pathway(GO:0097527) |
| 0.1 |
0.5 |
GO:0098734 |
protein depalmitoylation(GO:0002084) positive regulation of pinocytosis(GO:0048549) macromolecule depalmitoylation(GO:0098734) |
| 0.1 |
1.0 |
GO:1902018 |
negative regulation of cilium assembly(GO:1902018) |
| 0.1 |
0.4 |
GO:0032627 |
interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) |
| 0.1 |
0.2 |
GO:0016321 |
female meiosis chromosome segregation(GO:0016321) |
| 0.1 |
1.6 |
GO:0070208 |
protein heterotrimerization(GO:0070208) |
| 0.1 |
0.3 |
GO:0002677 |
negative regulation of chronic inflammatory response(GO:0002677) |
| 0.1 |
1.1 |
GO:0042640 |
anagen(GO:0042640) |
| 0.1 |
0.9 |
GO:0030150 |
protein import into mitochondrial matrix(GO:0030150) |
| 0.1 |
0.7 |
GO:1990173 |
protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.1 |
0.3 |
GO:0032532 |
regulation of microvillus length(GO:0032532) |
| 0.1 |
0.4 |
GO:0046477 |
glycosylceramide catabolic process(GO:0046477) |
| 0.1 |
2.0 |
GO:0000266 |
mitochondrial fission(GO:0000266) |
| 0.1 |
1.8 |
GO:0035456 |
response to interferon-beta(GO:0035456) |
| 0.0 |
0.1 |
GO:2000832 |
negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.0 |
0.1 |
GO:0006434 |
seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 |
0.4 |
GO:0007000 |
nucleolus organization(GO:0007000) |
| 0.0 |
0.5 |
GO:0000722 |
telomere maintenance via recombination(GO:0000722) |
| 0.0 |
0.2 |
GO:0050747 |
positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 |
0.7 |
GO:0046599 |
regulation of centriole replication(GO:0046599) |
| 0.0 |
2.9 |
GO:0061077 |
chaperone-mediated protein folding(GO:0061077) |
| 0.0 |
2.0 |
GO:0045600 |
positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 |
0.4 |
GO:0072010 |
renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 |
0.4 |
GO:0043011 |
myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 |
0.1 |
GO:0045014 |
detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.0 |
0.7 |
GO:0031571 |
mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 |
0.1 |
GO:0072531 |
pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.0 |
0.2 |
GO:0046415 |
urate metabolic process(GO:0046415) |
| 0.0 |
0.2 |
GO:0016081 |
synaptic vesicle docking(GO:0016081) |
| 0.0 |
0.1 |
GO:0060718 |
chorionic trophoblast cell differentiation(GO:0060718) |
| 0.0 |
0.4 |
GO:0006465 |
signal peptide processing(GO:0006465) |
| 0.0 |
0.9 |
GO:0046785 |
microtubule polymerization(GO:0046785) |
| 0.0 |
0.1 |
GO:0000479 |
endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |