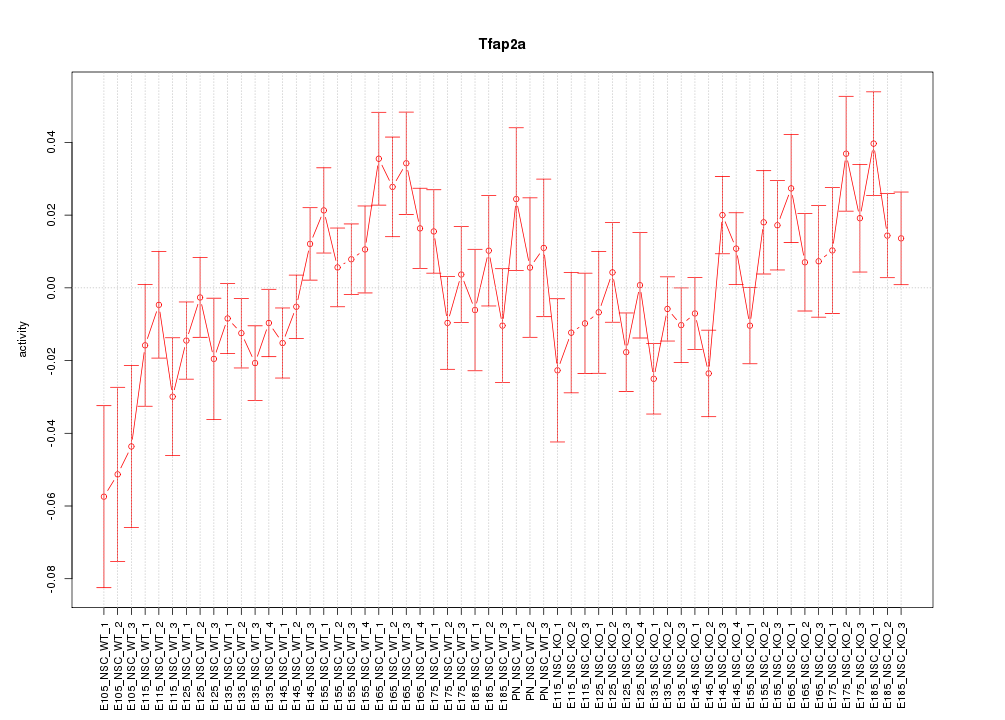
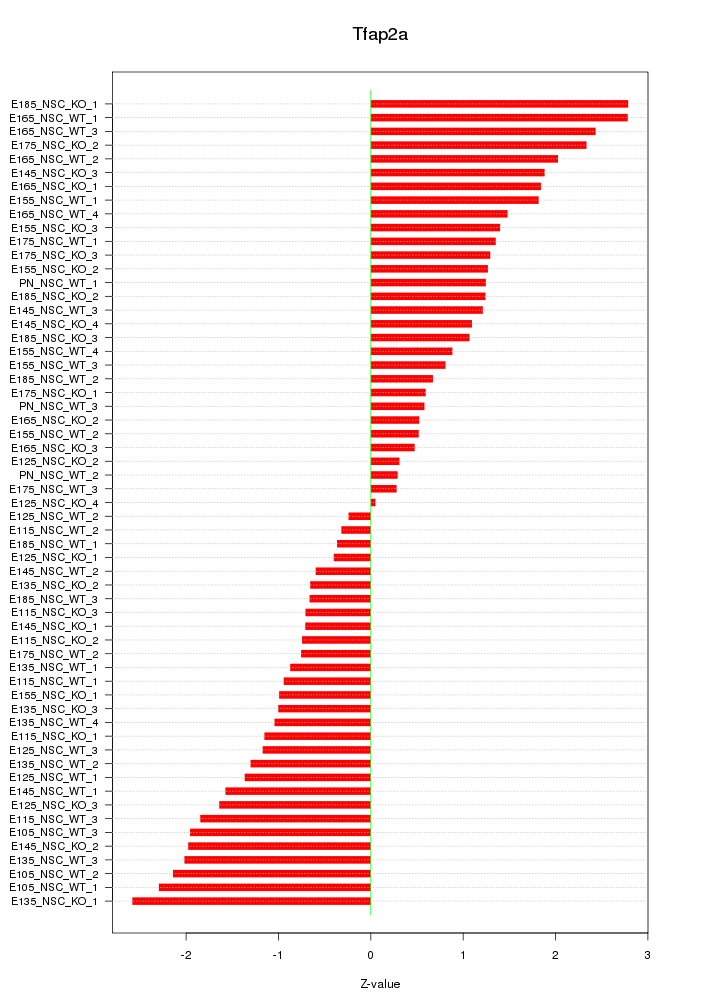
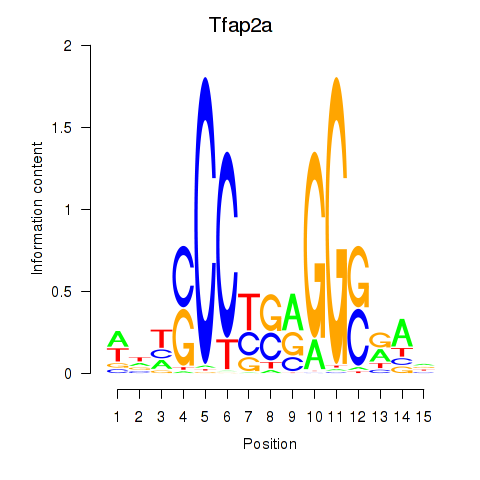
Motif ID: Tfap2a
Z-value: 1.382
Transcription factors associated with Tfap2a:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Tfap2a | ENSMUSG00000021359.9 | Tfap2a |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tfap2a | mm10_v2_chr13_-_40733768_40733836 | -0.36 | 5.3e-03 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 16.4 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 3.5 | 10.4 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 2.6 | 13.0 | GO:0010624 | regulation of Schwann cell proliferation(GO:0010624) |
| 2.4 | 30.7 | GO:0070842 | aggresome assembly(GO:0070842) |
| 2.3 | 7.0 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 2.2 | 8.6 | GO:1901204 | positive regulation of guanylate cyclase activity(GO:0031284) regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 2.0 | 8.0 | GO:0033024 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 1.7 | 5.1 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) ceramide transport(GO:0035627) |
| 1.7 | 8.3 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 1.4 | 4.3 | GO:0046959 | habituation(GO:0046959) |
| 1.4 | 5.8 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 1.3 | 6.7 | GO:0070305 | response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) |
| 1.3 | 3.8 | GO:0014877 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 1.3 | 6.3 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 1.2 | 5.9 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 1.1 | 4.4 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 1.1 | 6.6 | GO:0000820 | regulation of glutamine family amino acid metabolic process(GO:0000820) |
| 1.1 | 4.3 | GO:0032289 | central nervous system myelin formation(GO:0032289) cardiac cell fate specification(GO:0060912) |
| 1.1 | 8.4 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 1.0 | 20.6 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 1.0 | 3.0 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 1.0 | 6.0 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 1.0 | 5.0 | GO:0051012 | microtubule sliding(GO:0051012) negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 1.0 | 5.0 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 1.0 | 6.9 | GO:0032811 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) negative regulation of epinephrine secretion(GO:0032811) |
| 1.0 | 2.9 | GO:0071544 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.9 | 2.8 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.9 | 2.8 | GO:0021933 | radial glia guided migration of cerebellar granule cell(GO:0021933) |
| 0.9 | 2.7 | GO:0008228 | opsonization(GO:0008228) |
| 0.8 | 4.0 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.8 | 3.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.7 | 4.5 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.7 | 2.2 | GO:0061535 | glutamate secretion, neurotransmission(GO:0061535) |
| 0.7 | 2.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.7 | 3.4 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.7 | 6.6 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.6 | 2.6 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.6 | 2.5 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.6 | 4.9 | GO:0002488 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway(GO:0002488) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent(GO:0002489) |
| 0.6 | 6.3 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.6 | 5.8 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.6 | 1.7 | GO:0019401 | alditol biosynthetic process(GO:0019401) |
| 0.6 | 2.9 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.6 | 2.8 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.5 | 3.7 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.5 | 1.5 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.5 | 2.5 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.5 | 4.8 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.5 | 2.4 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.5 | 2.4 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.5 | 1.9 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.5 | 6.4 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.5 | 4.6 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.4 | 2.2 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.4 | 2.2 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.4 | 10.2 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.4 | 12.0 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.4 | 2.0 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.4 | 1.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.4 | 0.4 | GO:0061324 | canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901963) |
| 0.4 | 2.3 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.4 | 1.1 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.4 | 1.5 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.4 | 2.2 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.4 | 2.6 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.3 | 1.4 | GO:1904177 | regulation of adipose tissue development(GO:1904177) |
| 0.3 | 1.0 | GO:1990314 | pyrimidine-containing compound transmembrane transport(GO:0072531) cellular response to insulin-like growth factor stimulus(GO:1990314) |
| 0.3 | 1.4 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.3 | 3.4 | GO:1900112 | regulation of histone H3-K9 trimethylation(GO:1900112) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.3 | 2.4 | GO:1903551 | regulation of extracellular exosome assembly(GO:1903551) |
| 0.3 | 1.3 | GO:1904996 | diapedesis(GO:0050904) positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.3 | 1.6 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.3 | 1.0 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.3 | 1.3 | GO:0010046 | arginine biosynthetic process(GO:0006526) response to mycotoxin(GO:0010046) |
| 0.3 | 0.9 | GO:0036216 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.3 | 1.2 | GO:0010985 | negative regulation of lipoprotein particle clearance(GO:0010985) |
| 0.3 | 1.2 | GO:1903861 | regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.3 | 1.2 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.3 | 4.7 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.3 | 7.8 | GO:0095500 | acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.3 | 1.7 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.3 | 4.5 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.3 | 1.4 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.3 | 1.4 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.3 | 1.9 | GO:1903423 | positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.3 | 1.1 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.3 | 2.9 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.3 | 1.6 | GO:0030397 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.3 | 1.0 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.3 | 1.0 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.3 | 1.8 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.3 | 1.0 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.3 | 1.5 | GO:0098700 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.2 | 2.4 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.2 | 1.4 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.2 | 3.8 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.2 | 0.5 | GO:1903351 | response to dopamine(GO:1903350) cellular response to dopamine(GO:1903351) |
| 0.2 | 1.4 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.2 | 2.3 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.2 | 1.4 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.2 | 6.8 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.2 | 3.4 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.2 | 1.3 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.2 | 5.1 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.2 | 2.8 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.2 | 3.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.2 | 1.5 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.2 | 5.5 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.2 | 1.7 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.2 | 1.0 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.2 | 0.8 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.2 | 1.2 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.2 | 3.0 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.2 | 1.0 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.2 | 3.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.2 | 2.3 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.2 | 0.7 | GO:0006842 | tricarboxylic acid transport(GO:0006842) succinate transport(GO:0015744) citrate transport(GO:0015746) |
| 0.2 | 1.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.2 | 0.9 | GO:0060046 | regulation of acrosome reaction(GO:0060046) |
| 0.2 | 5.0 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.2 | 0.4 | GO:1904192 | cholangiocyte apoptotic process(GO:1902488) positive regulation of sensory perception of pain(GO:1904058) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 0.2 | 2.0 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.2 | 2.1 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.2 | 1.7 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.2 | 0.8 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.2 | 0.8 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.2 | 4.0 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.2 | 0.5 | GO:0033239 | negative regulation of cellular amine metabolic process(GO:0033239) |
| 0.2 | 1.0 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.2 | 0.5 | GO:0070625 | zymogen granule exocytosis(GO:0070625) |
| 0.2 | 1.8 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.2 | 4.1 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.1 | 2.7 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 0.9 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.1 | 0.4 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.1 | 1.0 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 | 4.4 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 0.5 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.1 | 2.6 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.1 | 0.3 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.1 | 0.4 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.1 | 1.3 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 1.4 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.1 | 2.5 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.1 | 0.7 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.1 | 0.8 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.1 | 0.9 | GO:0033005 | positive regulation of mast cell activation(GO:0033005) positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.1 | 2.5 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 0.3 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 | 0.5 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 6.7 | GO:0099643 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.1 | 0.2 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.1 | 0.4 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 | 2.1 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.1 | 3.0 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 0.3 | GO:0045414 | regulation of interleukin-8 biosynthetic process(GO:0045414) |
| 0.1 | 0.3 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.1 | 1.2 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 2.6 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 8.2 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.1 | 0.8 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 0.3 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 1.8 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.1 | 1.9 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.1 | 0.3 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.1 | 0.6 | GO:0099515 | actin filament-based transport(GO:0099515) |
| 0.1 | 0.9 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.1 | 0.6 | GO:0010640 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) |
| 0.1 | 0.5 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 2.9 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 0.7 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) sphingoid biosynthetic process(GO:0046520) |
| 0.1 | 0.5 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.1 | 3.6 | GO:0001938 | positive regulation of endothelial cell proliferation(GO:0001938) |
| 0.1 | 1.4 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.1 | 2.8 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 0.5 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 | 0.6 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.1 | 1.8 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 0.2 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 0.3 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 1.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 1.0 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) mRNA destabilization(GO:0061157) |
| 0.1 | 0.5 | GO:0008209 | androgen metabolic process(GO:0008209) |
| 0.1 | 0.7 | GO:0002643 | regulation of tolerance induction(GO:0002643) |
| 0.1 | 0.5 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 0.8 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 0.6 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.1 | 2.4 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 1.9 | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway(GO:0042058) |
| 0.1 | 2.8 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 3.3 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.1 | 2.1 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.1 | 0.5 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.1 | 6.3 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.1 | 2.2 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.1 | 0.6 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 0.5 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.1 | 6.2 | GO:0017157 | regulation of exocytosis(GO:0017157) |
| 0.1 | 1.1 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.1 | 0.4 | GO:0035729 | response to hepatocyte growth factor(GO:0035728) cellular response to hepatocyte growth factor stimulus(GO:0035729) |
| 0.1 | 0.3 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 1.4 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.0 | 2.3 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 2.5 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.0 | 0.7 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 1.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 3.9 | GO:0051924 | regulation of calcium ion transport(GO:0051924) |
| 0.0 | 0.2 | GO:0032796 | uropod organization(GO:0032796) natural killer cell degranulation(GO:0043320) |
| 0.0 | 1.0 | GO:2000772 | regulation of cellular senescence(GO:2000772) |
| 0.0 | 2.3 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 6.2 | GO:0006874 | cellular calcium ion homeostasis(GO:0006874) |
| 0.0 | 0.3 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 1.0 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.2 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.9 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.0 | 2.4 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 1.1 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 2.6 | GO:0006497 | protein lipidation(GO:0006497) |
| 0.0 | 0.9 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.0 | 0.9 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 1.7 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 0.1 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.0 | 0.1 | GO:0022410 | circadian sleep/wake cycle process(GO:0022410) regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) circadian sleep/wake cycle, sleep(GO:0050802) |
| 0.0 | 1.6 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 1.2 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.6 | GO:0019319 | hexose biosynthetic process(GO:0019319) |
| 0.0 | 1.9 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.2 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.2 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.5 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.0 | 1.0 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 1.0 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 1.1 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.6 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.6 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.4 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 2.0 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 0.3 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.0 | 0.7 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 2.3 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.3 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.1 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) very long-chain fatty acid biosynthetic process(GO:0042761) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 10.4 | GO:0072534 | perineuronal net(GO:0072534) |
| 3.3 | 13.0 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 1.5 | 4.5 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 1.4 | 6.9 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 1.1 | 4.4 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.9 | 15.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.9 | 2.8 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.8 | 4.0 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.8 | 3.9 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.7 | 7.9 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.7 | 2.7 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.6 | 3.9 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.6 | 8.3 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.5 | 4.8 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.5 | 2.6 | GO:0000798 | nuclear cohesin complex(GO:0000798) nuclear meiotic cohesin complex(GO:0034991) |
| 0.5 | 10.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.5 | 3.7 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.5 | 1.4 | GO:0005940 | septin ring(GO:0005940) |
| 0.4 | 1.3 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.4 | 1.7 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.3 | 1.0 | GO:0070522 | ERCC4-ERCC1 complex(GO:0070522) |
| 0.3 | 5.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.3 | 16.2 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.3 | 2.8 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.3 | 1.6 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.3 | 11.8 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.3 | 0.9 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.3 | 0.8 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.3 | 3.5 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.3 | 1.5 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.2 | 3.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 2.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.2 | 1.0 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.2 | 5.6 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.2 | 6.0 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 1.6 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.2 | 1.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.2 | 6.1 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.2 | 7.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.2 | 8.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.2 | 2.1 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.2 | 3.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.2 | 5.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.2 | 4.5 | GO:0043218 | compact myelin(GO:0043218) |
| 0.2 | 2.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.2 | 1.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.2 | 0.9 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.2 | 1.8 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 1.6 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.2 | 1.3 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.2 | 5.8 | GO:0032420 | stereocilium(GO:0032420) |
| 0.2 | 2.8 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.2 | 1.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.1 | 1.8 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.8 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 33.9 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 0.4 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.1 | 2.6 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 2.3 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.1 | 1.9 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 2.0 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 0.8 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 0.6 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 2.0 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.1 | 5.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 2.3 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.1 | 0.5 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 0.3 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.1 | 1.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 1.0 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 7.0 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 4.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 6.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 2.8 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 12.3 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 0.4 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 0.8 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 1.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 4.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 0.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 1.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 2.6 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 2.5 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 2.0 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 3.6 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 1.9 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.4 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 2.7 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 1.4 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 3.2 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 8.8 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 3.2 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.4 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.9 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 2.6 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.2 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 1.2 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 4.7 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 5.7 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.6 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.3 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 0.7 | GO:0097060 | synaptic membrane(GO:0097060) |
| 0.0 | 2.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 3.7 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 5.8 | GO:0097708 | cytoplasmic vesicle(GO:0031410) intracellular vesicle(GO:0097708) |
| 0.0 | 1.1 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.0 | 0.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 1.4 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.0 | GO:0071010 | prespliceosome(GO:0071010) |
| 0.0 | 9.6 | GO:0005794 | Golgi apparatus(GO:0005794) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 20.6 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 2.9 | 8.6 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 2.8 | 17.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 1.9 | 5.7 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 1.4 | 8.4 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 1.4 | 4.2 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 1.3 | 8.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 1.2 | 3.7 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 1.2 | 4.8 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 1.1 | 3.4 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 1.1 | 6.9 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 1.1 | 6.6 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 1.0 | 15.5 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 1.0 | 8.9 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.9 | 8.3 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.9 | 4.4 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.9 | 6.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.8 | 3.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.8 | 4.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.8 | 4.1 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.8 | 4.0 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.8 | 4.0 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.7 | 2.2 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.7 | 5.4 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.7 | 2.7 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.7 | 2.6 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.6 | 5.2 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.6 | 4.5 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.6 | 7.0 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.6 | 3.0 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.6 | 2.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.6 | 2.9 | GO:0018121 | imidazoleglycerol-phosphate synthase activity(GO:0000107) NAD(P)-cysteine ADP-ribosyltransferase activity(GO:0018071) NAD(P)-asparagine ADP-ribosyltransferase activity(GO:0018121) NAD(P)-serine ADP-ribosyltransferase activity(GO:0018127) 7-cyano-7-deazaguanine tRNA-ribosyltransferase activity(GO:0043867) purine deoxyribosyltransferase activity(GO:0044102) |
| 0.6 | 4.6 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.6 | 1.7 | GO:0008521 | acetyl-CoA transporter activity(GO:0008521) |
| 0.5 | 2.7 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.5 | 2.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.5 | 2.6 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.5 | 1.5 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.5 | 1.0 | GO:0046978 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.5 | 1.5 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.5 | 2.9 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.5 | 1.4 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.5 | 1.4 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.5 | 4.1 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.4 | 1.3 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.4 | 3.9 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.4 | 1.7 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.4 | 1.2 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.4 | 2.0 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.4 | 1.6 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.4 | 1.5 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.4 | 8.7 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.4 | 1.8 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) |
| 0.4 | 2.5 | GO:0016937 | short-branched-chain-acyl-CoA dehydrogenase activity(GO:0016937) |
| 0.4 | 0.7 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.3 | 2.4 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.3 | 1.4 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.3 | 2.0 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.3 | 1.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.3 | 2.3 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.3 | 1.5 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.3 | 1.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.3 | 2.3 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.3 | 2.0 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.3 | 5.2 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.3 | 1.3 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.3 | 3.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.3 | 3.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.2 | 0.7 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.2 | 2.4 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.2 | 3.6 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.2 | 0.7 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.2 | 0.5 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.2 | 1.4 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.2 | 3.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.2 | 0.6 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.2 | 0.8 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.2 | 1.4 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.2 | 7.5 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.2 | 2.2 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.2 | 1.0 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.2 | 1.7 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.2 | 1.5 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.2 | 1.4 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 3.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.2 | 5.0 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.2 | 2.4 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.2 | 6.7 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.2 | 0.5 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.2 | 8.9 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.2 | 1.6 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.2 | 6.1 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.2 | 0.5 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) |
| 0.2 | 5.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.2 | 1.6 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.1 | 10.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 2.5 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.6 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 1.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 3.3 | GO:0071617 | lysophospholipid acyltransferase activity(GO:0071617) |
| 0.1 | 2.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 2.7 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 1.1 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 1.0 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 0.6 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 0.3 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.1 | 2.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 2.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 1.0 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 2.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 16.3 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 0.8 | GO:0046703 | MHC class I protein binding(GO:0042288) natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.1 | 2.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 1.3 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 2.8 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.1 | 0.4 | GO:0080084 | RNA polymerase III type 1 promoter DNA binding(GO:0001030) RNA polymerase III type 2 promoter DNA binding(GO:0001031) RNA polymerase III type 3 promoter DNA binding(GO:0001032) 5S rDNA binding(GO:0080084) |
| 0.1 | 1.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 2.8 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 3.6 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.1 | 2.2 | GO:0004120 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) photoreceptor cyclic-nucleotide phosphodiesterase activity(GO:0004120) 7,8-dihydro-D-neopterin 2',3'-cyclic phosphate phosphodiesterase activity(GO:0044688) inositol phosphosphingolipid phospholipase activity(GO:0052712) inositol phosphorylceramide phospholipase activity(GO:0052713) mannosyl-inositol phosphorylceramide phospholipase activity(GO:0052714) mannosyl-diinositol phosphorylceramide phospholipase activity(GO:0052715) |
| 0.1 | 1.7 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 1.3 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 1.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.4 | GO:0042979 | ornithine decarboxylase regulator activity(GO:0042979) |
| 0.1 | 2.9 | GO:0008748 | N-ethylmaleimide reductase activity(GO:0008748) reduced coenzyme F420 dehydrogenase activity(GO:0043738) sulfur oxygenase reductase activity(GO:0043826) malolactic enzyme activity(GO:0043883) epoxyqueuosine reductase activity(GO:0052693) |
| 0.1 | 0.8 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 1.0 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 4.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 2.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 1.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 1.4 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.3 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 0.4 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 3.1 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.1 | 3.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 4.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.7 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 1.5 | GO:0052771 | coenzyme F390-A hydrolase activity(GO:0052770) coenzyme F390-G hydrolase activity(GO:0052771) |
| 0.1 | 0.3 | GO:0043842 | Kdo transferase activity(GO:0043842) |
| 0.0 | 9.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 2.4 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 1.5 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.5 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 1.0 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.4 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.5 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 2.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.2 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 9.5 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 1.8 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.2 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.7 | GO:0015605 | organophosphate ester transmembrane transporter activity(GO:0015605) |
| 0.0 | 0.1 | GO:1904680 | peptide transmembrane transporter activity(GO:1904680) |
| 0.0 | 0.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 2.1 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.3 | GO:0043028 | cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.8 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 3.3 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.8 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 4.0 | GO:0008514 | organic anion transmembrane transporter activity(GO:0008514) |
| 0.0 | 1.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.0 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) |
| 0.0 | 0.6 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.1 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.0 | 3.0 | GO:0003729 | mRNA binding(GO:0003729) |
| 0.0 | 0.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.8 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 3.7 | GO:0016757 | transferase activity, transferring glycosyl groups(GO:0016757) |
| 0.0 | 7.5 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 0.5 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.9 | GO:0101005 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.1 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |