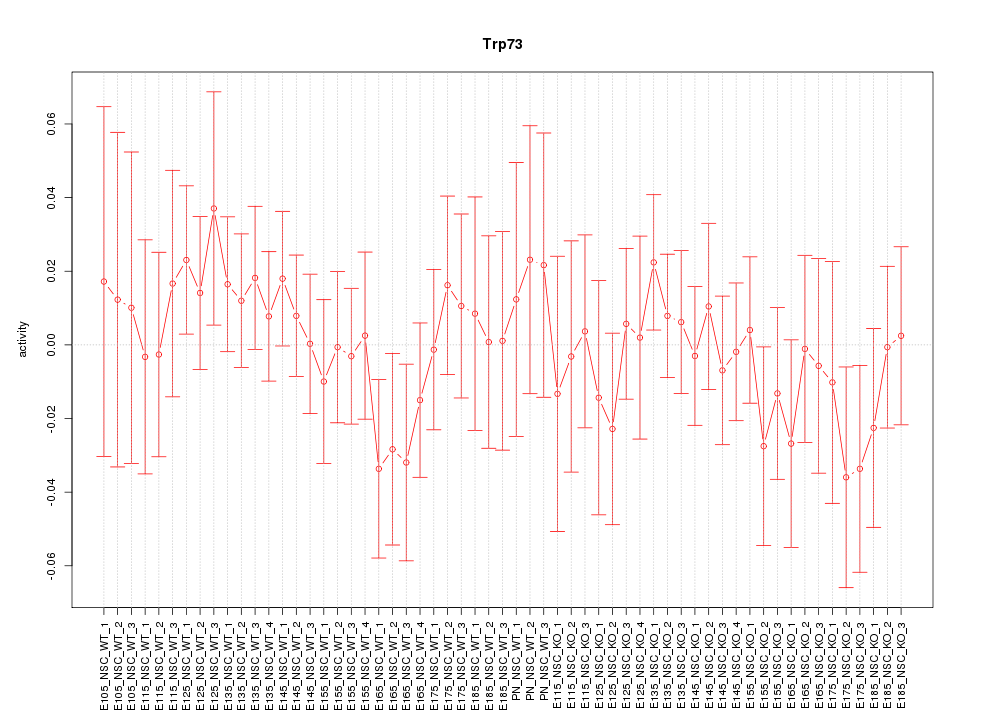
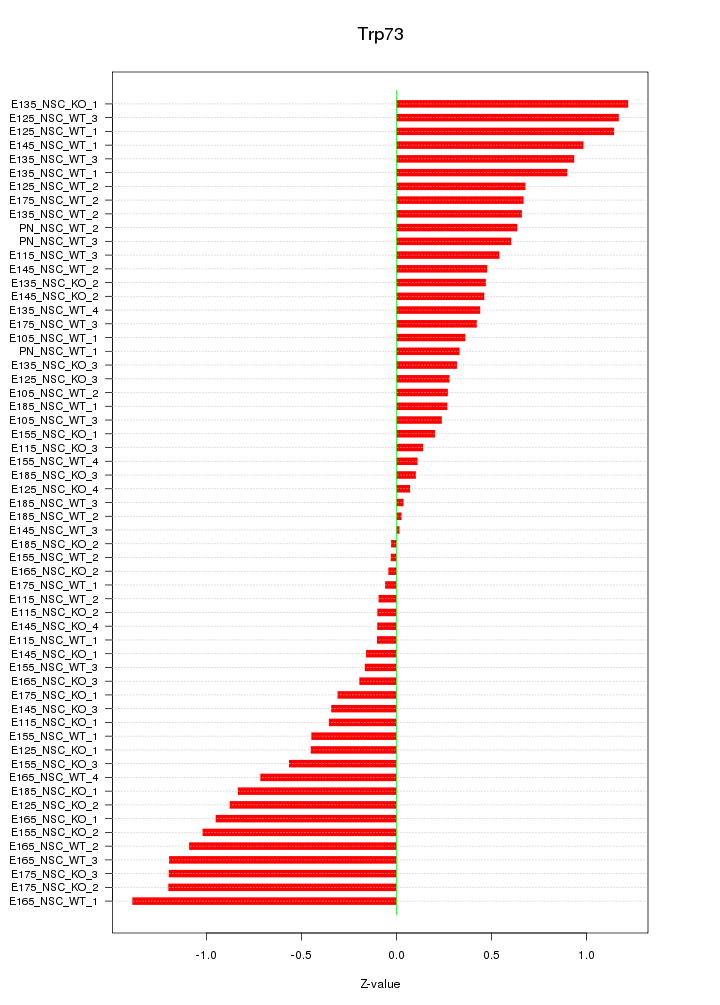
Motif ID: Trp73
Z-value: 0.631

Transcription factors associated with Trp73:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Trp73 | ENSMUSG00000029026.10 | Trp73 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Trp73 | mm10_v2_chr4_-_154097105_154097173 | 0.23 | 8.0e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.4 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 1.1 | 4.4 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.7 | 5.5 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.4 | 1.3 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.4 | 2.6 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.4 | 4.3 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.3 | 0.9 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.2 | 0.7 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.2 | 2.6 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.2 | 0.7 | GO:0002295 | T-helper cell lineage commitment(GO:0002295) interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.2 | 1.8 | GO:0071493 | cellular response to UV-B(GO:0071493) replicative senescence(GO:0090399) |
| 0.2 | 0.6 | GO:1903795 | regulation of inorganic anion transmembrane transport(GO:1903795) |
| 0.2 | 2.2 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.2 | 0.5 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.2 | 0.7 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.2 | 1.8 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.2 | 2.3 | GO:0042104 | positive regulation of activated T cell proliferation(GO:0042104) |
| 0.1 | 0.5 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.1 | 1.1 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.1 | 1.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.1 | 3.1 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.1 | 1.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.6 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.5 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 1.5 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 0.8 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.0 | 1.0 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.0 | 0.5 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 1.3 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.5 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.1 | GO:0035811 | negative regulation of urine volume(GO:0035811) |
| 0.0 | 0.8 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 1.2 | GO:0042035 | regulation of cytokine biosynthetic process(GO:0042035) |
| 0.0 | 1.1 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 1.0 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.5 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.1 | GO:0036120 | cellular response to platelet-derived growth factor stimulus(GO:0036120) positive regulation of tyrosine phosphorylation of Stat5 protein(GO:0042523) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0044299 | C-fiber(GO:0044299) |
| 0.6 | 1.8 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.4 | 1.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.3 | 5.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.2 | 2.6 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.2 | 1.4 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.8 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 2.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 0.6 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.8 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 2.0 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 0.7 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 1.0 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.5 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.9 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 1.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.7 | GO:0045120 | telomerase holoenzyme complex(GO:0005697) pronucleus(GO:0045120) |
| 0.0 | 0.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.1 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.5 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.4 | 1.2 | GO:0005118 | sevenless binding(GO:0005118) |
| 0.3 | 2.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 5.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.2 | 1.0 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.2 | 2.6 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.2 | 1.8 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.1 | 0.5 | GO:0004954 | icosanoid receptor activity(GO:0004953) prostanoid receptor activity(GO:0004954) prostaglandin receptor activity(GO:0004955) |
| 0.1 | 1.8 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.7 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.1 | 1.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 1.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.8 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.5 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 2.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 1.1 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.5 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.0 | 1.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.8 | GO:0034946 | 2-oxoglutaryl-CoA thioesterase activity(GO:0034843) 2,4,4-trimethyl-3-oxopentanoyl-CoA thioesterase activity(GO:0034869) 3-isopropylbut-3-enoyl-CoA thioesterase activity(GO:0034946) glutaryl-CoA hydrolase activity(GO:0044466) |
| 0.0 | 0.8 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 1.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 4.7 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.7 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.1 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.0 | 0.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.5 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 4.2 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.7 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 1.3 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.5 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 5.7 | GO:0046982 | protein heterodimerization activity(GO:0046982) |