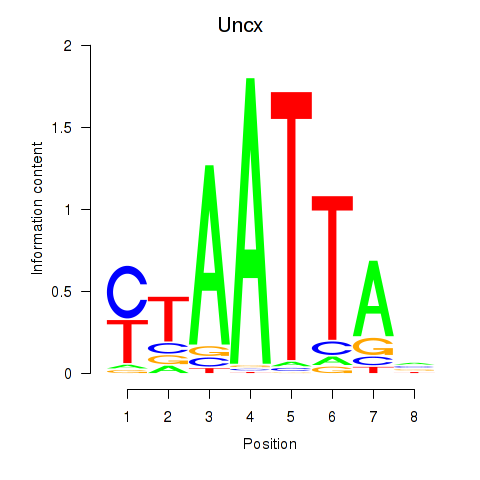
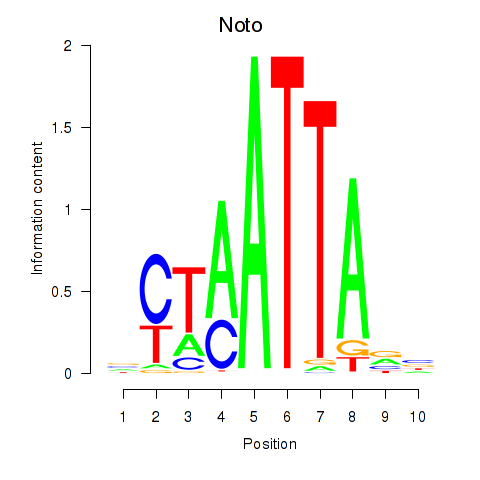
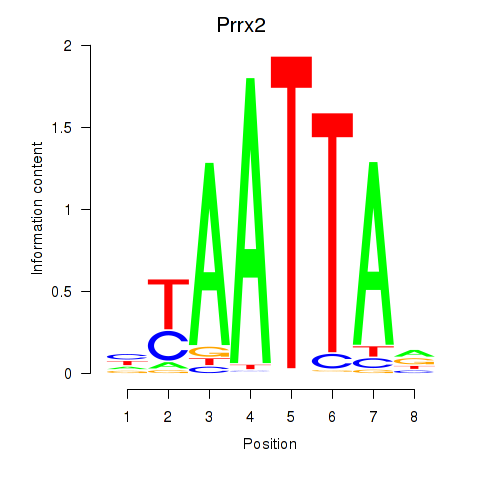
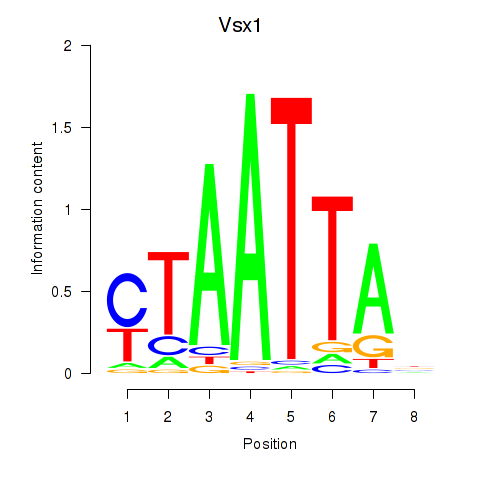
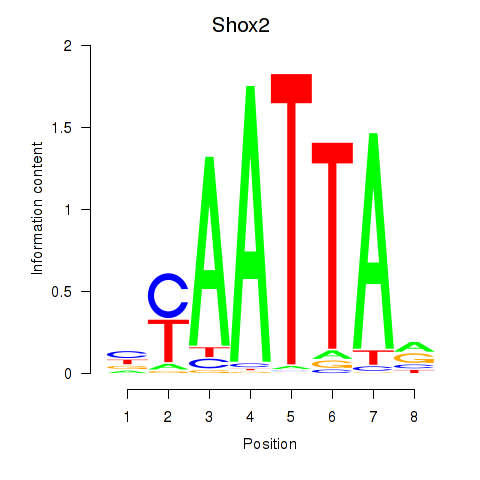
| 11.0 |
44.2 |
GO:0046552 |
eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 7.0 |
21.0 |
GO:0048818 |
embryonic nail plate morphogenesis(GO:0035880) positive regulation of hair follicle maturation(GO:0048818) positive regulation of catagen(GO:0051795) frontal suture morphogenesis(GO:0060364) |
| 4.7 |
14.2 |
GO:0048389 |
BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) intermediate mesoderm development(GO:0048389) regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) pattern specification involved in mesonephros development(GO:0061227) regulation of hepatocyte differentiation(GO:0070366) comma-shaped body morphogenesis(GO:0072049) S-shaped body morphogenesis(GO:0072050) anterior/posterior pattern specification involved in kidney development(GO:0072098) negative regulation of glomerular mesangial cell proliferation(GO:0072125) ureter urothelium development(GO:0072190) negative regulation of glomerulus development(GO:0090194) regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 3.2 |
25.8 |
GO:0035726 |
common myeloid progenitor cell proliferation(GO:0035726) |
| 2.5 |
7.6 |
GO:0072204 |
cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 2.4 |
43.5 |
GO:0001833 |
inner cell mass cell proliferation(GO:0001833) |
| 2.2 |
17.8 |
GO:0033353 |
S-adenosylmethionine cycle(GO:0033353) |
| 1.9 |
3.8 |
GO:0060741 |
prostate gland stromal morphogenesis(GO:0060741) |
| 1.9 |
7.5 |
GO:0021847 |
ventricular zone neuroblast division(GO:0021847) |
| 1.8 |
10.9 |
GO:0060648 |
mammary gland bud morphogenesis(GO:0060648) |
| 1.8 |
16.2 |
GO:0010587 |
miRNA catabolic process(GO:0010587) |
| 1.7 |
5.0 |
GO:0090425 |
hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 1.6 |
4.8 |
GO:0030916 |
otic vesicle formation(GO:0030916) |
| 1.6 |
20.6 |
GO:0031017 |
exocrine pancreas development(GO:0031017) |
| 1.6 |
4.7 |
GO:0003162 |
atrioventricular node development(GO:0003162) |
| 1.5 |
3.0 |
GO:0021529 |
spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 1.3 |
3.8 |
GO:0060166 |
olfactory pit development(GO:0060166) |
| 1.2 |
4.8 |
GO:1904565 |
response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 1.2 |
7.1 |
GO:0045218 |
zonula adherens maintenance(GO:0045218) |
| 1.2 |
3.5 |
GO:0051798 |
positive regulation of hair follicle development(GO:0051798) |
| 1.2 |
4.7 |
GO:0035553 |
oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 1.1 |
17.5 |
GO:0021521 |
ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 1.1 |
16.2 |
GO:0071803 |
positive regulation of podosome assembly(GO:0071803) |
| 1.0 |
3.1 |
GO:0042663 |
regulation of endodermal cell fate specification(GO:0042663) |
| 1.0 |
3.1 |
GO:0090324 |
negative regulation of oxidative phosphorylation(GO:0090324) |
| 1.0 |
3.9 |
GO:0021780 |
oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.9 |
15.1 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.9 |
4.4 |
GO:0007195 |
adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.9 |
1.7 |
GO:0042536 |
negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.8 |
2.4 |
GO:0015889 |
cobalamin transport(GO:0015889) |
| 0.8 |
4.8 |
GO:0036233 |
glycine import(GO:0036233) |
| 0.8 |
10.1 |
GO:0060159 |
regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.8 |
2.3 |
GO:0042320 |
regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) circadian sleep/wake cycle, REM sleep(GO:0042747) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.7 |
2.2 |
GO:1905065 |
positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 0.7 |
4.3 |
GO:0006123 |
mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.7 |
2.2 |
GO:0072071 |
apoptotic process involved in heart morphogenesis(GO:0003278) regulation of vascular wound healing(GO:0061043) mesangial cell differentiation(GO:0072007) glomerular mesangial cell differentiation(GO:0072008) kidney interstitial fibroblast differentiation(GO:0072071) renal interstitial fibroblast development(GO:0072141) mesangial cell development(GO:0072143) glomerular mesangial cell development(GO:0072144) |
| 0.7 |
3.6 |
GO:0007223 |
Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.7 |
4.2 |
GO:0009249 |
protein lipoylation(GO:0009249) |
| 0.7 |
1.3 |
GO:2000974 |
negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.6 |
1.9 |
GO:2000852 |
regulation of corticosterone secretion(GO:2000852) |
| 0.6 |
1.2 |
GO:1904956 |
regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.5 |
0.5 |
GO:0061743 |
motor learning(GO:0061743) |
| 0.5 |
1.5 |
GO:0090526 |
response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.5 |
12.7 |
GO:0009954 |
proximal/distal pattern formation(GO:0009954) |
| 0.4 |
2.8 |
GO:0034047 |
regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.4 |
11.6 |
GO:0006270 |
DNA replication initiation(GO:0006270) |
| 0.4 |
1.2 |
GO:0003221 |
right ventricular cardiac muscle tissue morphogenesis(GO:0003221) ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.4 |
1.9 |
GO:0006348 |
chromatin silencing at telomere(GO:0006348) |
| 0.4 |
1.9 |
GO:0015692 |
vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.4 |
2.3 |
GO:0070562 |
regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.4 |
6.3 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.4 |
1.4 |
GO:0000707 |
meiotic DNA recombinase assembly(GO:0000707) resolution of recombination intermediates(GO:0071139) |
| 0.3 |
2.8 |
GO:1902916 |
positive regulation of protein polyubiquitination(GO:1902916) |
| 0.3 |
5.7 |
GO:0021542 |
dentate gyrus development(GO:0021542) |
| 0.3 |
8.0 |
GO:0002089 |
lens morphogenesis in camera-type eye(GO:0002089) |
| 0.3 |
2.3 |
GO:0032049 |
cardiolipin biosynthetic process(GO:0032049) |
| 0.3 |
1.0 |
GO:0043586 |
tongue development(GO:0043586) |
| 0.3 |
0.6 |
GO:0060059 |
embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.3 |
0.6 |
GO:0043490 |
malate-aspartate shuttle(GO:0043490) |
| 0.3 |
2.7 |
GO:0045602 |
negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.3 |
2.7 |
GO:0060501 |
positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.3 |
3.2 |
GO:0061302 |
smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.3 |
1.5 |
GO:0006564 |
L-serine biosynthetic process(GO:0006564) |
| 0.3 |
0.9 |
GO:0042271 |
susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.3 |
1.7 |
GO:0042795 |
snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.3 |
4.1 |
GO:0007530 |
sex determination(GO:0007530) |
| 0.3 |
1.7 |
GO:0052695 |
cellular glucuronidation(GO:0052695) |
| 0.3 |
1.9 |
GO:0010908 |
regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.3 |
1.1 |
GO:0002326 |
B cell lineage commitment(GO:0002326) |
| 0.3 |
6.4 |
GO:0097352 |
autophagosome maturation(GO:0097352) |
| 0.2 |
1.0 |
GO:0001550 |
ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.2 |
5.3 |
GO:0045662 |
negative regulation of myoblast differentiation(GO:0045662) |
| 0.2 |
3.3 |
GO:0030497 |
fatty acid elongation(GO:0030497) |
| 0.2 |
0.7 |
GO:0006933 |
negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.2 |
2.1 |
GO:0043922 |
negative regulation by host of viral transcription(GO:0043922) |
| 0.2 |
0.7 |
GO:0050748 |
negative regulation of receptor biosynthetic process(GO:0010871) negative regulation of lipoprotein metabolic process(GO:0050748) |
| 0.2 |
3.4 |
GO:0006107 |
oxaloacetate metabolic process(GO:0006107) |
| 0.2 |
0.9 |
GO:0090365 |
regulation of mRNA modification(GO:0090365) |
| 0.2 |
11.2 |
GO:0042475 |
odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.2 |
0.7 |
GO:0035633 |
maintenance of blood-brain barrier(GO:0035633) |
| 0.2 |
1.5 |
GO:0007406 |
negative regulation of neuroblast proliferation(GO:0007406) |
| 0.2 |
10.6 |
GO:0051693 |
actin filament capping(GO:0051693) |
| 0.2 |
0.7 |
GO:0008228 |
opsonization(GO:0008228) modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.2 |
3.5 |
GO:0050860 |
negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.2 |
1.7 |
GO:0006003 |
fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 |
0.6 |
GO:0030210 |
heparin biosynthetic process(GO:0030210) Tie signaling pathway(GO:0048014) |
| 0.2 |
1.9 |
GO:0051764 |
actin crosslink formation(GO:0051764) |
| 0.2 |
1.0 |
GO:0038032 |
termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.2 |
1.2 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.2 |
1.4 |
GO:0032782 |
urea transport(GO:0015840) bile acid secretion(GO:0032782) urea transmembrane transport(GO:0071918) |
| 0.2 |
5.6 |
GO:0018345 |
protein palmitoylation(GO:0018345) |
| 0.2 |
0.4 |
GO:1903977 |
positive regulation of Schwann cell migration(GO:1900149) positive regulation of glial cell migration(GO:1903977) |
| 0.2 |
3.3 |
GO:0042541 |
hemoglobin biosynthetic process(GO:0042541) |
| 0.2 |
0.5 |
GO:0090271 |
positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.2 |
1.0 |
GO:0016078 |
tRNA catabolic process(GO:0016078) |
| 0.2 |
0.3 |
GO:0060023 |
soft palate development(GO:0060023) |
| 0.2 |
1.4 |
GO:0045199 |
maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.2 |
1.0 |
GO:0021756 |
striatum development(GO:0021756) |
| 0.2 |
0.8 |
GO:0010835 |
regulation of protein ADP-ribosylation(GO:0010835) |
| 0.1 |
3.3 |
GO:0035335 |
peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 |
0.7 |
GO:0010359 |
regulation of anion channel activity(GO:0010359) |
| 0.1 |
5.2 |
GO:0045494 |
photoreceptor cell maintenance(GO:0045494) |
| 0.1 |
1.7 |
GO:0019985 |
translesion synthesis(GO:0019985) |
| 0.1 |
3.1 |
GO:0007638 |
mechanosensory behavior(GO:0007638) |
| 0.1 |
3.6 |
GO:0030901 |
midbrain development(GO:0030901) |
| 0.1 |
0.5 |
GO:1900740 |
regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.1 |
0.6 |
GO:0040032 |
post-embryonic body morphogenesis(GO:0040032) hair follicle placode formation(GO:0060789) |
| 0.1 |
1.2 |
GO:1903504 |
regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 |
3.5 |
GO:0031365 |
N-terminal protein amino acid modification(GO:0031365) |
| 0.1 |
0.9 |
GO:0060355 |
positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.1 |
0.8 |
GO:0031848 |
protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 |
1.9 |
GO:1902043 |
positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.1 |
0.8 |
GO:0071763 |
nuclear membrane organization(GO:0071763) |
| 0.1 |
0.4 |
GO:0016259 |
selenocysteine metabolic process(GO:0016259) |
| 0.1 |
1.0 |
GO:0080111 |
DNA demethylation(GO:0080111) |
| 0.1 |
0.7 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 |
0.4 |
GO:0034285 |
response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.1 |
1.7 |
GO:0006999 |
nuclear pore organization(GO:0006999) |
| 0.1 |
1.1 |
GO:0099625 |
regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) ventricular cardiac muscle cell membrane repolarization(GO:0099625) |
| 0.1 |
3.6 |
GO:0007029 |
endoplasmic reticulum organization(GO:0007029) |
| 0.1 |
1.7 |
GO:0010172 |
embryonic body morphogenesis(GO:0010172) |
| 0.1 |
0.6 |
GO:0035095 |
behavioral response to nicotine(GO:0035095) |
| 0.1 |
1.6 |
GO:0033194 |
response to hydroperoxide(GO:0033194) |
| 0.1 |
5.6 |
GO:0051965 |
positive regulation of synapse assembly(GO:0051965) |
| 0.1 |
0.7 |
GO:0009396 |
folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.1 |
1.7 |
GO:0000132 |
establishment of mitotic spindle orientation(GO:0000132) |
| 0.1 |
0.5 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 |
1.7 |
GO:0050873 |
brown fat cell differentiation(GO:0050873) |
| 0.1 |
1.5 |
GO:0007026 |
negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 |
7.8 |
GO:0030178 |
negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.1 |
0.2 |
GO:2000822 |
regulation of fear response(GO:1903365) positive regulation of fear response(GO:1903367) regulation of behavioral fear response(GO:2000822) positive regulation of behavioral fear response(GO:2000987) |
| 0.1 |
0.2 |
GO:0015712 |
hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) phosphate ion transmembrane transport(GO:0035435) |
| 0.1 |
0.9 |
GO:0008340 |
determination of adult lifespan(GO:0008340) |
| 0.1 |
4.4 |
GO:0048706 |
embryonic skeletal system development(GO:0048706) |
| 0.0 |
1.4 |
GO:0006006 |
glucose metabolic process(GO:0006006) |
| 0.0 |
0.5 |
GO:0046548 |
retinal rod cell development(GO:0046548) |
| 0.0 |
2.5 |
GO:0002224 |
toll-like receptor signaling pathway(GO:0002224) |
| 0.0 |
0.7 |
GO:0035136 |
forelimb morphogenesis(GO:0035136) |
| 0.0 |
0.7 |
GO:0003351 |
epithelial cilium movement(GO:0003351) |
| 0.0 |
0.2 |
GO:0051697 |
protein delipidation(GO:0051697) |
| 0.0 |
2.7 |
GO:0030168 |
platelet activation(GO:0030168) |
| 0.0 |
0.3 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.0 |
1.1 |
GO:0007566 |
embryo implantation(GO:0007566) |
| 0.0 |
2.6 |
GO:0007601 |
visual perception(GO:0007601) |
| 0.0 |
0.9 |
GO:0071384 |
cellular response to corticosteroid stimulus(GO:0071384) cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 |
0.1 |
GO:0042420 |
dopamine catabolic process(GO:0042420) |
| 0.0 |
2.2 |
GO:0050728 |
negative regulation of inflammatory response(GO:0050728) |
| 0.0 |
0.7 |
GO:0071108 |
protein K48-linked deubiquitination(GO:0071108) |
| 0.0 |
0.3 |
GO:0032225 |
regulation of synaptic transmission, dopaminergic(GO:0032225) |
| 0.0 |
1.7 |
GO:0006367 |
transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 |
0.7 |
GO:0006491 |
N-glycan processing(GO:0006491) |
| 0.0 |
0.6 |
GO:0002053 |
positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.0 |
0.4 |
GO:0016973 |
poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 |
0.6 |
GO:0006101 |
tricarboxylic acid cycle(GO:0006099) citrate metabolic process(GO:0006101) |
| 0.0 |
9.7 |
GO:0007067 |
mitotic nuclear division(GO:0007067) |
| 0.0 |
2.3 |
GO:0035023 |
regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 |
0.6 |
GO:0009060 |
aerobic respiration(GO:0009060) |
| 0.0 |
3.2 |
GO:0008360 |
regulation of cell shape(GO:0008360) |
| 0.0 |
0.7 |
GO:0006284 |
base-excision repair(GO:0006284) |
| 0.0 |
0.2 |
GO:0031987 |
locomotion involved in locomotory behavior(GO:0031987) |
| 0.0 |
1.5 |
GO:0051028 |
mRNA transport(GO:0051028) |
| 0.0 |
0.2 |
GO:0006953 |
acute-phase response(GO:0006953) |
| 0.0 |
6.7 |
GO:0008284 |
positive regulation of cell proliferation(GO:0008284) |
| 0.0 |
0.4 |
GO:0006940 |
regulation of smooth muscle contraction(GO:0006940) |
| 0.0 |
1.9 |
GO:0071383 |
cellular response to steroid hormone stimulus(GO:0071383) |
| 0.0 |
2.3 |
GO:1902593 |
protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) nuclear import(GO:0051170) single-organism nuclear import(GO:1902593) |
| 0.0 |
0.1 |
GO:1903352 |
ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.0 |
1.0 |
GO:0019882 |
antigen processing and presentation(GO:0019882) |
| 0.0 |
0.3 |
GO:0043984 |
histone H4-K16 acetylation(GO:0043984) |
| 0.0 |
0.2 |
GO:0007189 |
adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 |
0.3 |
GO:0000154 |
rRNA modification(GO:0000154) |
| 0.0 |
0.3 |
GO:0032922 |
circadian regulation of gene expression(GO:0032922) |