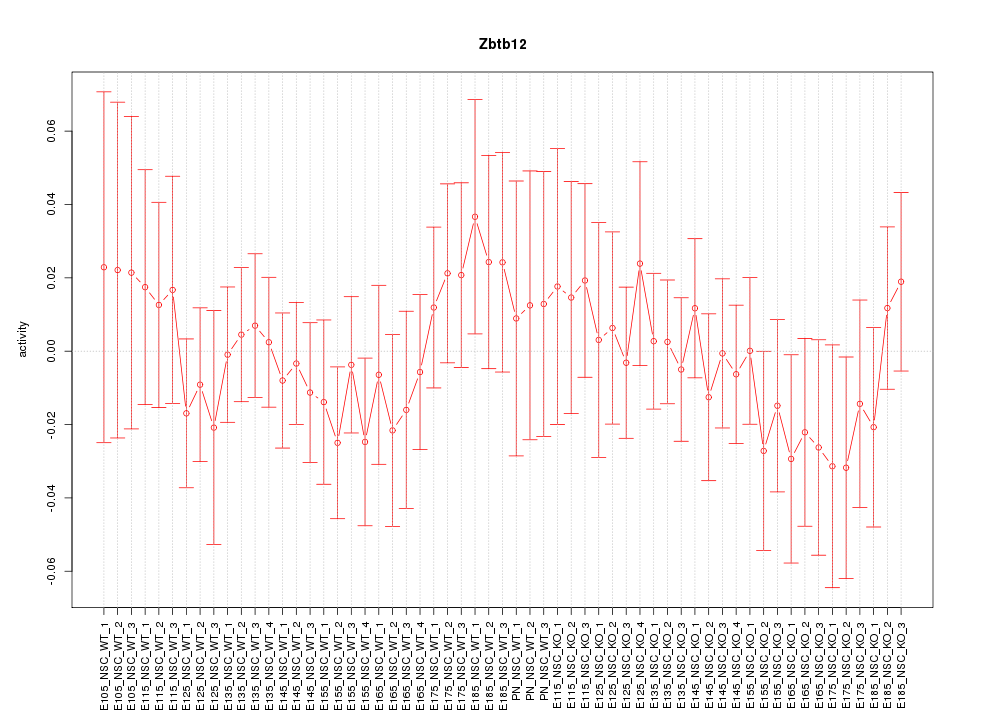
| 1.3 |
3.9 |
GO:0019405 |
alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 1.2 |
3.6 |
GO:0014826 |
vein smooth muscle contraction(GO:0014826) |
| 1.2 |
3.5 |
GO:0010248 |
B cell negative selection(GO:0002352) establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 1.0 |
3.1 |
GO:0060853 |
arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) endothelial cell fate specification(GO:0060847) Notch signaling pathway involved in arterial endothelial cell fate commitment(GO:0060853) blood vessel endothelial cell fate specification(GO:0097101) |
| 0.6 |
1.8 |
GO:1900275 |
negative regulation of phospholipase C activity(GO:1900275) |
| 0.5 |
1.6 |
GO:0018094 |
protein polyglycylation(GO:0018094) |
| 0.5 |
2.9 |
GO:0098700 |
aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.5 |
4.4 |
GO:0045719 |
negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.4 |
3.3 |
GO:0033601 |
positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.4 |
1.2 |
GO:0009174 |
UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.4 |
1.1 |
GO:0002586 |
positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.4 |
1.8 |
GO:0051012 |
microtubule sliding(GO:0051012) |
| 0.3 |
0.9 |
GO:0033634 |
positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.3 |
6.1 |
GO:0042744 |
hydrogen peroxide catabolic process(GO:0042744) |
| 0.3 |
2.4 |
GO:0032286 |
central nervous system myelin maintenance(GO:0032286) |
| 0.2 |
1.9 |
GO:0002154 |
thyroid hormone mediated signaling pathway(GO:0002154) regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.2 |
3.3 |
GO:0035493 |
SNARE complex assembly(GO:0035493) |
| 0.2 |
0.5 |
GO:1903896 |
positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.1 |
2.9 |
GO:0005980 |
polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 |
0.6 |
GO:0070562 |
regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 |
0.7 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 |
1.3 |
GO:0072010 |
renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.1 |
1.3 |
GO:0050860 |
negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 |
2.3 |
GO:0032008 |
positive regulation of TOR signaling(GO:0032008) |
| 0.1 |
0.2 |
GO:0070682 |
proteasome regulatory particle assembly(GO:0070682) |
| 0.0 |
0.2 |
GO:1901301 |
regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 |
0.0 |
GO:1901187 |
regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 |
2.0 |
GO:0030901 |
midbrain development(GO:0030901) |
| 0.0 |
0.8 |
GO:0034063 |
stress granule assembly(GO:0034063) |
| 0.0 |
0.4 |
GO:0070286 |
axonemal dynein complex assembly(GO:0070286) |
| 0.0 |
4.2 |
GO:0007050 |
cell cycle arrest(GO:0007050) |
| 0.0 |
0.9 |
GO:0030514 |
negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 |
0.9 |
GO:0007032 |
endosome organization(GO:0007032) |
| 0.0 |
3.6 |
GO:0051321 |
meiotic cell cycle(GO:0051321) |
| 0.0 |
0.2 |
GO:0042407 |
cristae formation(GO:0042407) |
| 0.0 |
0.2 |
GO:0043923 |
positive regulation by host of viral transcription(GO:0043923) |
| 0.0 |
0.3 |
GO:0071364 |
cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 |
2.5 |
GO:0048864 |
stem cell development(GO:0048864) |
| 0.0 |
0.0 |
GO:0006049 |
UDP-N-acetylglucosamine catabolic process(GO:0006049) |