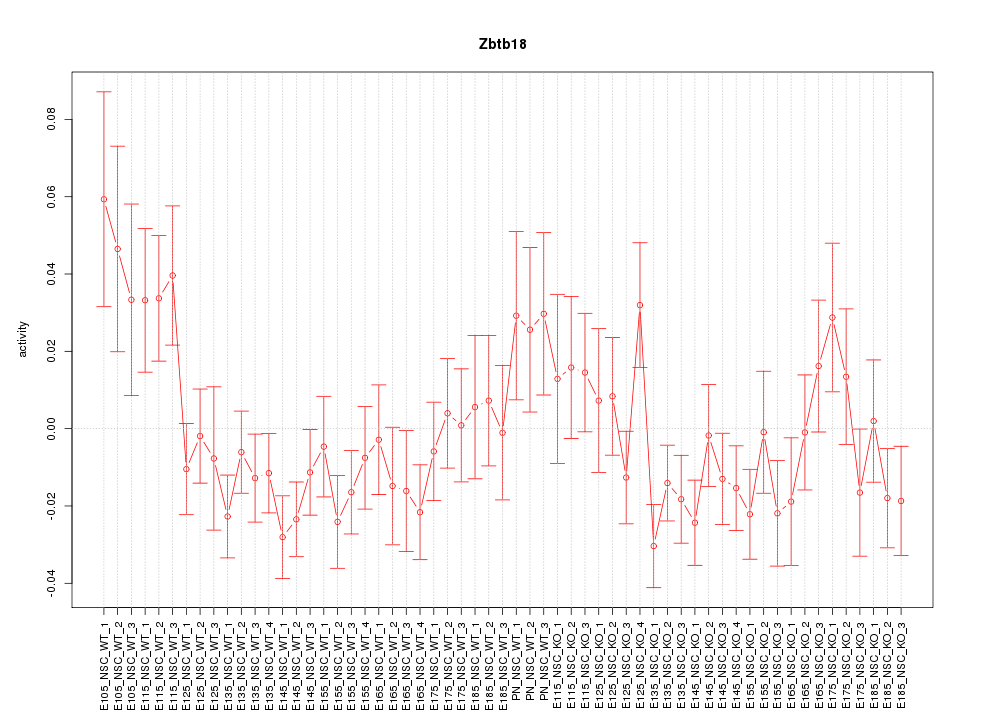
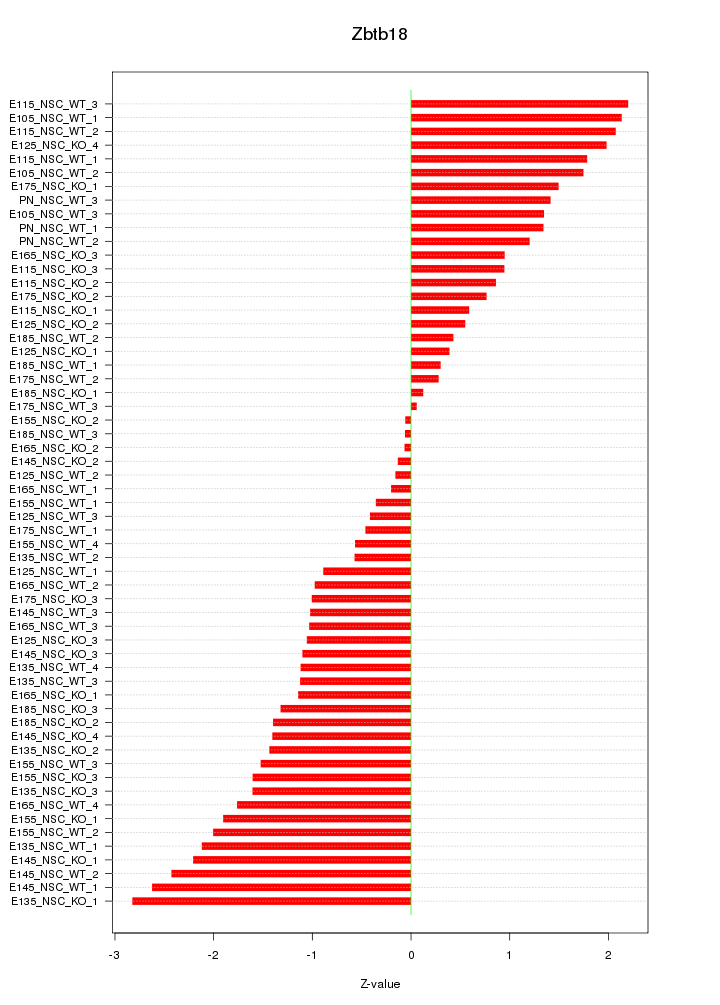
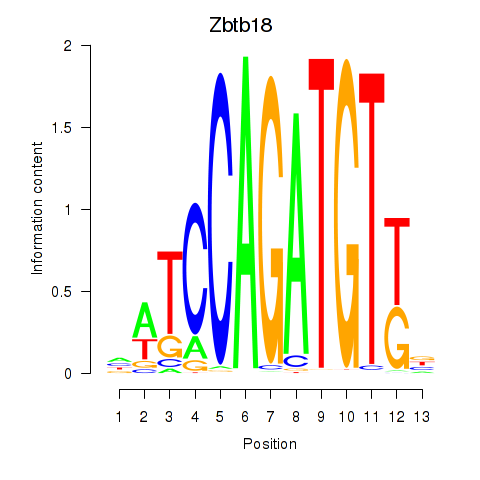
Motif ID: Zbtb18
Z-value: 1.339
Transcription factors associated with Zbtb18:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Zbtb18 | ENSMUSG00000063659.6 | Zbtb18 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb18 | mm10_v2_chr1_+_177445660_177445821 | -0.75 | 9.9e-12 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 20.0 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 3.0 | 9.0 | GO:0060769 | positive regulation of epithelial cell proliferation involved in prostate gland development(GO:0060769) kidney smooth muscle tissue development(GO:0072194) negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 2.1 | 6.4 | GO:0045404 | positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 2.0 | 6.0 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 1.8 | 5.4 | GO:2001139 | negative regulation of postsynaptic membrane organization(GO:1901627) negative regulation of dendritic spine maintenance(GO:1902951) negative regulation of phospholipid efflux(GO:1902999) regulation of lipid transport across blood brain barrier(GO:1903000) negative regulation of lipid transport across blood brain barrier(GO:1903001) positive regulation of lipid transport across blood brain barrier(GO:1903002) negative regulation of phospholipid transport(GO:2001139) |
| 1.5 | 4.4 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 1.5 | 4.4 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 1.1 | 3.4 | GO:0060364 | frontal suture morphogenesis(GO:0060364) |
| 1.1 | 3.4 | GO:0030300 | regulation of intestinal cholesterol absorption(GO:0030300) |
| 1.0 | 4.2 | GO:0048539 | bone marrow development(GO:0048539) |
| 1.0 | 3.1 | GO:0034162 | toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.9 | 2.8 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.9 | 0.9 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.9 | 3.6 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 0.9 | 4.4 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.8 | 2.5 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.8 | 2.4 | GO:2000852 | regulation of corticosterone secretion(GO:2000852) |
| 0.7 | 3.0 | GO:0071033 | nuclear retention of pre-mRNA at the site of transcription(GO:0071033) CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.7 | 2.2 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.7 | 3.6 | GO:1903012 | positive regulation of bone development(GO:1903012) |
| 0.7 | 7.0 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.7 | 1.4 | GO:0061344 | regulation of cell adhesion involved in heart morphogenesis(GO:0061344) positive regulation of ephrin receptor signaling pathway(GO:1901189) |
| 0.7 | 4.6 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.7 | 5.9 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.6 | 1.9 | GO:0010757 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) negative regulation of plasminogen activation(GO:0010757) regulation of vascular wound healing(GO:0061043) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.6 | 1.9 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.6 | 3.0 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.6 | 1.8 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.6 | 13.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.6 | 1.7 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.6 | 2.3 | GO:0046210 | nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.6 | 1.7 | GO:0006059 | hexitol metabolic process(GO:0006059) alditol biosynthetic process(GO:0019401) |
| 0.6 | 2.3 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.6 | 2.2 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.5 | 1.6 | GO:0001803 | antibody-dependent cellular cytotoxicity(GO:0001788) type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 0.5 | 2.1 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.5 | 2.0 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.5 | 2.0 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.5 | 1.9 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.5 | 4.3 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.5 | 3.6 | GO:0060290 | transdifferentiation(GO:0060290) |
| 0.4 | 1.7 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.4 | 2.2 | GO:0019659 | lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.4 | 4.2 | GO:0046036 | GTP biosynthetic process(GO:0006183) CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.4 | 2.1 | GO:0046864 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) alveolar primary septum development(GO:0061143) |
| 0.4 | 2.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.4 | 2.7 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.4 | 1.5 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.4 | 1.1 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.4 | 1.9 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.4 | 1.1 | GO:0040009 | regulation of growth rate(GO:0040009) protein poly-ADP-ribosylation(GO:0070212) |
| 0.4 | 1.4 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.4 | 1.4 | GO:1904451 | regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.4 | 2.8 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.3 | 3.8 | GO:0035814 | negative regulation of renal sodium excretion(GO:0035814) |
| 0.3 | 4.9 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.3 | 1.0 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.3 | 3.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.3 | 2.0 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.3 | 6.8 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.3 | 1.3 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.3 | 2.2 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.3 | 1.5 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.3 | 0.6 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.3 | 5.8 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.3 | 2.6 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.3 | 2.6 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.3 | 1.2 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.3 | 1.6 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.3 | 0.8 | GO:2000314 | negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.3 | 3.4 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.3 | 3.4 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.3 | 2.6 | GO:0009186 | deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.3 | 1.0 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.3 | 0.8 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.3 | 0.8 | GO:0002295 | T-helper cell lineage commitment(GO:0002295) |
| 0.2 | 0.7 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.2 | 1.5 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.2 | 4.3 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.2 | 1.9 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.2 | 4.4 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.2 | 2.7 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.2 | 1.3 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.2 | 2.0 | GO:0090037 | positive regulation of protein kinase C signaling(GO:0090037) |
| 0.2 | 3.6 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.2 | 2.1 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.2 | 2.3 | GO:1990173 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.2 | 1.0 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.2 | 0.6 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.2 | 1.4 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.2 | 2.8 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.2 | 1.0 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.2 | 3.7 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.2 | 1.9 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.2 | 1.7 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.2 | 1.3 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.2 | 0.9 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.2 | 1.5 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.2 | 0.5 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.2 | 0.5 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.2 | 0.5 | GO:0035811 | negative regulation of urine volume(GO:0035811) |
| 0.2 | 0.6 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.2 | 2.7 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.2 | 1.4 | GO:0097531 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) mast cell migration(GO:0097531) |
| 0.2 | 6.8 | GO:0032611 | interleukin-1 beta production(GO:0032611) |
| 0.2 | 0.5 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 1.8 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.1 | 4.1 | GO:0030201 | heparan sulfate proteoglycan metabolic process(GO:0030201) |
| 0.1 | 0.6 | GO:0015819 | lysine transport(GO:0015819) |
| 0.1 | 0.8 | GO:0070813 | hydrogen sulfide metabolic process(GO:0070813) |
| 0.1 | 1.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.1 | 5.2 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.1 | 0.8 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.1 | 1.7 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.1 | 0.5 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 0.4 | GO:0097278 | complement-dependent cytotoxicity(GO:0097278) |
| 0.1 | 3.9 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 0.8 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.1 | 4.7 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 | 2.8 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 5.6 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.1 | 1.1 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.6 | GO:2000252 | progesterone secretion(GO:0042701) negative regulation of feeding behavior(GO:2000252) |
| 0.1 | 2.1 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.1 | 0.7 | GO:1902993 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.1 | 0.8 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.1 | 4.4 | GO:0033014 | porphyrin-containing compound biosynthetic process(GO:0006779) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.1 | 0.3 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 1.2 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 0.5 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 | 0.5 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.1 | 0.4 | GO:0070589 | cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) cell wall biogenesis(GO:0042546) cell wall macromolecule metabolic process(GO:0044036) cell wall macromolecule biosynthetic process(GO:0044038) chain elongation of O-linked mannose residue(GO:0044845) cellular component macromolecule biosynthetic process(GO:0070589) cell wall organization or biogenesis(GO:0071554) |
| 0.1 | 1.8 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 1.3 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 1.6 | GO:0046596 | regulation of viral entry into host cell(GO:0046596) |
| 0.1 | 0.7 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.7 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 0.3 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.1 | 1.4 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 | 0.9 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.4 | GO:0061525 | hindgut development(GO:0061525) |
| 0.1 | 2.3 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.1 | 2.9 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.1 | 0.4 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 1.3 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.1 | 0.6 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 2.1 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.1 | 0.8 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 2.8 | GO:0048678 | response to axon injury(GO:0048678) |
| 0.1 | 2.4 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 4.7 | GO:0034620 | cellular response to unfolded protein(GO:0034620) |
| 0.1 | 0.4 | GO:0048149 | adult feeding behavior(GO:0008343) behavioral response to ethanol(GO:0048149) |
| 0.1 | 0.5 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.1 | 0.2 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 1.4 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.1 | 0.2 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.1 | 0.4 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 2.0 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.1 | 0.6 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 0.4 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.1 | 0.2 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.1 | 1.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 3.4 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 0.7 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 3.4 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.0 | 2.9 | GO:0048663 | neuron fate commitment(GO:0048663) |
| 0.0 | 1.0 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.4 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.2 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.0 | 0.1 | GO:0015960 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.0 | 1.1 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 1.8 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 2.4 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 0.1 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.0 | 2.0 | GO:0001942 | hair follicle development(GO:0001942) molting cycle process(GO:0022404) hair cycle process(GO:0022405) skin epidermis development(GO:0098773) |
| 0.0 | 3.6 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 0.6 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.9 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.0 | 0.5 | GO:0046628 | positive regulation of insulin receptor signaling pathway(GO:0046628) |
| 0.0 | 0.3 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 4.1 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 0.5 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.0 | 4.0 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 1.1 | GO:0070613 | regulation of protein processing(GO:0070613) |
| 0.0 | 0.1 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 0.2 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 1.4 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 2.3 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.3 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.3 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.0 | 0.1 | GO:0060548 | negative regulation of cell death(GO:0060548) |
| 0.0 | 0.7 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 1.2 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.3 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 1.0 | GO:0060828 | regulation of canonical Wnt signaling pathway(GO:0060828) |
| 0.0 | 0.2 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 1.5 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.0 | 0.2 | GO:1901409 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 2.0 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.3 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.0 | 0.2 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) regulation of glucose import in response to insulin stimulus(GO:2001273) |
| 0.0 | 1.3 | GO:0000956 | nuclear-transcribed mRNA catabolic process(GO:0000956) |
| 0.0 | 1.5 | GO:0000082 | G1/S transition of mitotic cell cycle(GO:0000082) |
| 0.0 | 1.0 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.8 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 1.1 | GO:0045765 | regulation of angiogenesis(GO:0045765) |
| 0.0 | 0.3 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 0.9 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 | 1.0 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.4 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 1.2 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.6 | GO:0031047 | gene silencing by RNA(GO:0031047) |
| 0.0 | 1.0 | GO:0021537 | telencephalon development(GO:0021537) |
| 0.0 | 1.4 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.6 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.5 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.0 | 0.9 | GO:0045216 | cell-cell junction organization(GO:0045216) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.4 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 1.8 | 7.0 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 1.5 | 20.0 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 1.3 | 3.8 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.8 | 2.4 | GO:0036125 | mitochondrial fatty acid beta-oxidation multienzyme complex(GO:0016507) fatty acid beta-oxidation multienzyme complex(GO:0036125) |
| 0.7 | 2.2 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.7 | 2.7 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.6 | 1.9 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.5 | 1.6 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.5 | 2.6 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.4 | 1.3 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.4 | 1.7 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.4 | 1.9 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.4 | 1.4 | GO:1990795 | lateral part of cell(GO:0097574) basolateral part of cell(GO:1990794) rod bipolar cell terminal bouton(GO:1990795) |
| 0.4 | 1.1 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.3 | 1.4 | GO:0090537 | CERF complex(GO:0090537) |
| 0.3 | 1.0 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.3 | 3.4 | GO:0098533 | ATPase dependent transmembrane transport complex(GO:0098533) |
| 0.3 | 1.0 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.3 | 0.9 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.3 | 2.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.3 | 0.9 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.3 | 1.4 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.3 | 5.1 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.3 | 3.6 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.3 | 5.9 | GO:0005605 | basal lamina(GO:0005605) |
| 0.3 | 1.4 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.3 | 0.8 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.2 | 1.7 | GO:0030677 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.2 | 0.7 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.2 | 1.3 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.2 | 1.7 | GO:0032009 | early phagosome(GO:0032009) |
| 0.2 | 3.0 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.2 | 2.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.2 | 1.5 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.2 | 2.4 | GO:0032797 | SMN complex(GO:0032797) |
| 0.2 | 2.1 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.2 | 1.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.2 | 1.0 | GO:0036157 | axonemal dynein complex(GO:0005858) outer dynein arm(GO:0036157) |
| 0.2 | 1.0 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.2 | 2.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.2 | 0.5 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 1.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 3.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 10.7 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 1.1 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 1.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 0.4 | GO:0070449 | elongin complex(GO:0070449) |
| 0.1 | 1.9 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.1 | 0.5 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 8.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 0.8 | GO:0001740 | Barr body(GO:0001740) |
| 0.1 | 1.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.4 | GO:0031502 | dolichyl-phosphate-mannose-protein mannosyltransferase complex(GO:0031502) |
| 0.1 | 7.4 | GO:0005657 | replication fork(GO:0005657) |
| 0.1 | 4.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 0.8 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 1.4 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 1.0 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 0.6 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 4.3 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.1 | 0.3 | GO:0002111 | BRCA2-BRAF35 complex(GO:0002111) |
| 0.1 | 11.1 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 14.5 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 3.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 3.2 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.4 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 0.5 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 4.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 10.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 2.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 0.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 1.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 3.5 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 3.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.3 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.3 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.9 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 5.9 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.1 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 1.7 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.5 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.4 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 0.6 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 1.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 2.1 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.2 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.5 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.9 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.6 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.5 | GO:0036477 | somatodendritic compartment(GO:0036477) |
| 0.0 | 1.5 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 3.7 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 0.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 2.6 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.4 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 0.6 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 2.3 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 2.9 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 1.1 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 4.0 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 4.5 | GO:0019866 | organelle inner membrane(GO:0019866) |
| 0.0 | 0.4 | GO:0015030 | Cajal body(GO:0015030) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 14.9 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 1.8 | 5.4 | GO:0046911 | hydroxyapatite binding(GO:0046848) metal chelating activity(GO:0046911) phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 1.5 | 4.4 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 1.5 | 4.4 | GO:0005534 | galactose binding(GO:0005534) |
| 1.5 | 4.4 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 1.4 | 5.6 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 1.3 | 20.0 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.9 | 2.8 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.9 | 4.3 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.8 | 13.2 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.8 | 2.3 | GO:0005119 | smoothened binding(GO:0005119) hedgehog receptor activity(GO:0008158) hedgehog family protein binding(GO:0097108) |
| 0.8 | 6.0 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.7 | 2.2 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.6 | 2.4 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.6 | 1.8 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.6 | 1.7 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.6 | 2.3 | GO:0016708 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) iron-cytochrome-c reductase activity(GO:0047726) |
| 0.5 | 1.6 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.5 | 2.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.5 | 2.6 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.5 | 1.8 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.4 | 2.7 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.4 | 4.0 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.4 | 1.3 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.4 | 1.9 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.4 | 2.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.3 | 1.0 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.3 | 4.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.3 | 2.0 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.3 | 1.6 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.3 | 1.9 | GO:0052630 | CTP:2,3-di-O-geranylgeranyl-sn-glycero-1-phosphate cytidyltransferase activity(GO:0043338) phospholactate guanylyltransferase activity(GO:0043814) ATP:coenzyme F420 adenylyltransferase activity(GO:0043910) UDP-N-acetylgalactosamine diphosphorylase activity(GO:0052630) |
| 0.3 | 3.4 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.3 | 2.1 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.3 | 3.4 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.3 | 1.8 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.3 | 2.4 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.3 | 4.1 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.3 | 1.2 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.3 | 3.6 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.3 | 4.7 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.3 | 1.5 | GO:0034916 | 4-methyloctanoyl-CoA dehydrogenase activity(GO:0034580) naphthyl-2-methyl-succinyl-CoA dehydrogenase activity(GO:0034845) 2-methylhexanoyl-CoA dehydrogenase activity(GO:0034916) propionyl-CoA dehydrogenase activity(GO:0043820) thiol-driven fumarate reductase activity(GO:0043830) coenzyme F420-dependent 2,4,6-trinitrophenol reductase activity(GO:0052758) coenzyme F420-dependent 2,4,6-trinitrophenol hydride reductase activity(GO:0052759) coenzyme F420-dependent 2,4-dinitrophenol reductase activity(GO:0052760) |
| 0.3 | 1.5 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.2 | 0.7 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.2 | 1.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.2 | 0.9 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.2 | 2.8 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.2 | 0.9 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.2 | 4.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.2 | 9.0 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.2 | 2.4 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.2 | 1.5 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.2 | 0.6 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.2 | 1.4 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.2 | 2.8 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.2 | 1.6 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.2 | 3.1 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.2 | 2.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.2 | 0.7 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.2 | 1.1 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.2 | 2.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.2 | 3.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 1.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.2 | 1.1 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.2 | 1.7 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.2 | 0.5 | GO:0046997 | sarcosine dehydrogenase activity(GO:0008480) oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.2 | 4.7 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.2 | 1.1 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.1 | 1.1 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.1 | 2.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 0.5 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.5 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.4 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.1 | 5.0 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 1.0 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.1 | 1.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 1.7 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 3.7 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 1.5 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.6 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 1.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 2.3 | GO:0034946 | 2-oxoglutaryl-CoA thioesterase activity(GO:0034843) 2,4,4-trimethyl-3-oxopentanoyl-CoA thioesterase activity(GO:0034869) 3-isopropylbut-3-enoyl-CoA thioesterase activity(GO:0034946) glutaryl-CoA hydrolase activity(GO:0044466) |
| 0.1 | 1.7 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.1 | 1.0 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 1.6 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 0.8 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 2.0 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 3.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 2.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 0.6 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 1.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 1.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 3.4 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 0.4 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.1 | 1.9 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 0.7 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 2.7 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.1 | 0.5 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 0.6 | GO:0015189 | arginine transmembrane transporter activity(GO:0015181) L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 1.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.5 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.1 | 0.5 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 2.8 | GO:0008748 | N-ethylmaleimide reductase activity(GO:0008748) reduced coenzyme F420 dehydrogenase activity(GO:0043738) sulfur oxygenase reductase activity(GO:0043826) malolactic enzyme activity(GO:0043883) epoxyqueuosine reductase activity(GO:0052693) |
| 0.1 | 2.8 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 0.8 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 0.3 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.4 | GO:0002135 | CTP binding(GO:0002135) |
| 0.1 | 1.7 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 0.7 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.4 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 1.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 1.6 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.1 | 4.9 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 0.2 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 1.7 | GO:0034945 | dihydrolipoamide branched chain acyltransferase activity(GO:0004147) palmitoleoyl [acyl-carrier-protein]-dependent acyltransferase activity(GO:0008951) serine O-acyltransferase activity(GO:0016412) O-succinyltransferase activity(GO:0016750) sinapoyltransferase activity(GO:0016752) O-sinapoyltransferase activity(GO:0016753) peptidyl-lysine N6-myristoyltransferase activity(GO:0018030) peptidyl-lysine N6-palmitoyltransferase activity(GO:0018031) benzoyl acetate-CoA thiolase activity(GO:0018711) 3-hydroxybutyryl-CoA thiolase activity(GO:0018712) 3-ketopimelyl-CoA thiolase activity(GO:0018713) N-palmitoyltransferase activity(GO:0019105) acyl-CoA N-acyltransferase activity(GO:0019186) protein-cysteine S-myristoyltransferase activity(GO:0019705) glucosaminyl-phosphotidylinositol O-acyltransferase activity(GO:0032216) ergosterol O-acyltransferase activity(GO:0034737) lanosterol O-acyltransferase activity(GO:0034738) naphthyl-2-oxomethyl-succinyl-CoA succinyl transferase activity(GO:0034848) 2,4,4-trimethyl-3-oxopentanoyl-CoA 2-C-propanoyl transferase activity(GO:0034851) 2-methylhexanoyl-CoA C-acetyltransferase activity(GO:0034915) butyryl-CoA 2-C-propionyltransferase activity(GO:0034919) 2,6-dimethyl-5-methylene-3-oxo-heptanoyl-CoA C-acetyltransferase activity(GO:0034945) L-2-aminoadipate N-acetyltransferase activity(GO:0043741) keto acid formate lyase activity(GO:0043806) azetidine-2-carboxylic acid acetyltransferase activity(GO:0046941) peptidyl-lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0052858) acetyl-CoA:L-lysine N6-acetyltransferase(GO:0090595) |
| 0.0 | 3.2 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 2.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 1.7 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.3 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 10.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.0 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 3.9 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.7 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.8 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 2.1 | GO:0044824 | integrase activity(GO:0008907) T/G mismatch-specific endonuclease activity(GO:0043765) retroviral integrase activity(GO:0044823) retroviral 3' processing activity(GO:0044824) |
| 0.0 | 1.5 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 1.4 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 4.0 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 1.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.4 | GO:0034951 | pivalyl-CoA mutase activity(GO:0034784) o-hydroxylaminobenzoate mutase activity(GO:0034951) lupeol synthase activity(GO:0042299) beta-amyrin synthase activity(GO:0042300) baruol synthase activity(GO:0080011) |
| 0.0 | 1.5 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 1.4 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 7.3 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.3 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 3.2 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.3 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.8 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 0.5 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 7.2 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.4 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 1.1 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 2.5 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.9 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.9 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.6 | GO:0045502 | dynein binding(GO:0045502) |
| 0.0 | 0.4 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 1.1 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.6 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.7 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.8 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.3 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.6 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |