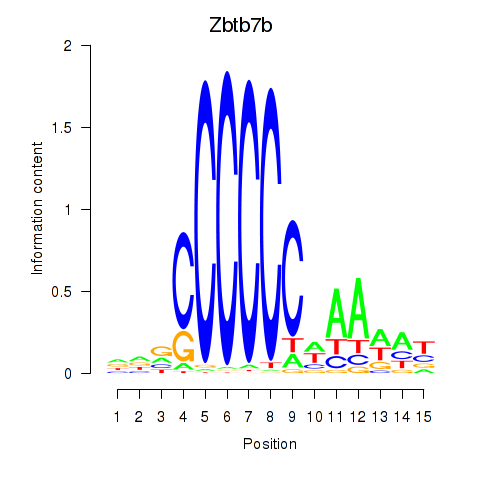
Motif ID: Zbtb7b
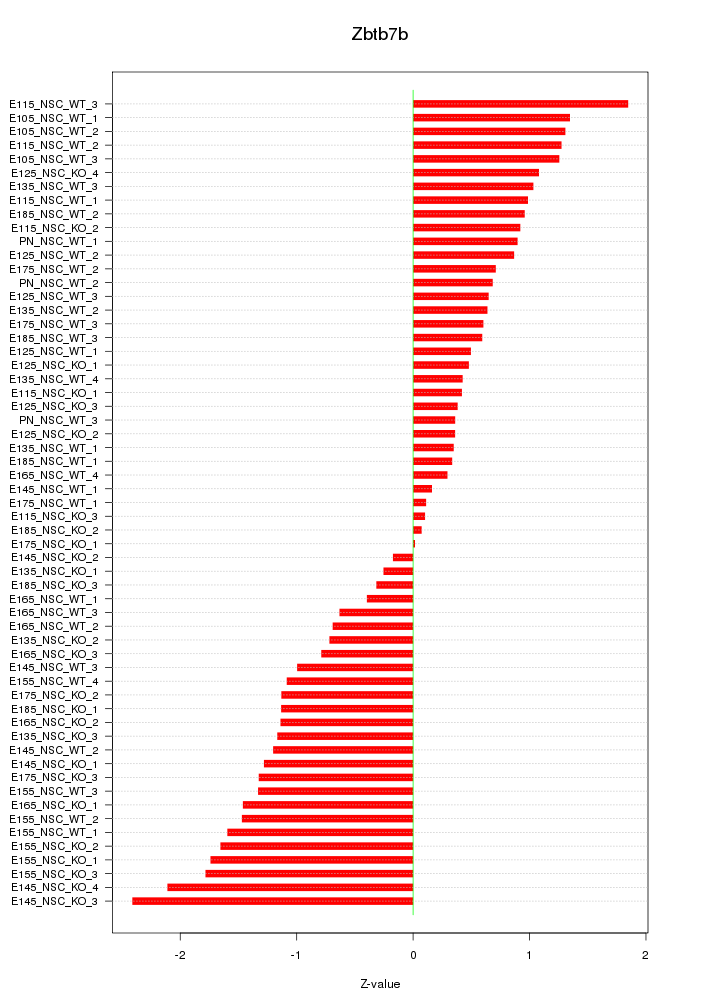
Z-value: 1.035
Transcription factors associated with Zbtb7b:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Zbtb7b | ENSMUSG00000028042.9 | Zbtb7b |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb7b | mm10_v2_chr3_-_89393294_89393378 | 0.40 | 1.7e-03 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 11.3 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 2.5 | 7.5 | GO:0097402 | neuroblast migration(GO:0097402) |
| 1.8 | 5.3 | GO:0048818 | embryonic nail plate morphogenesis(GO:0035880) positive regulation of hair follicle maturation(GO:0048818) positive regulation of catagen(GO:0051795) frontal suture morphogenesis(GO:0060364) |
| 1.7 | 8.6 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 1.6 | 6.3 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 1.6 | 6.3 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 1.5 | 13.3 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 1.3 | 3.8 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 1.2 | 3.6 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 1.1 | 9.6 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 1.1 | 3.2 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 1.0 | 4.0 | GO:0060032 | notochord regression(GO:0060032) |
| 1.0 | 4.0 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) lens fiber cell apoptotic process(GO:1990086) |
| 0.9 | 7.4 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.9 | 9.0 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.8 | 2.4 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.7 | 2.2 | GO:0048352 | neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) |
| 0.7 | 5.0 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.7 | 4.6 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.6 | 4.8 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.6 | 3.5 | GO:2001185 | regulation of CD8-positive, alpha-beta T cell activation(GO:2001185) |
| 0.6 | 4.1 | GO:0072539 | T-helper 17 cell differentiation(GO:0072539) |
| 0.6 | 1.7 | GO:1903795 | regulation of inorganic anion transmembrane transport(GO:1903795) |
| 0.5 | 1.6 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.5 | 3.5 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.5 | 3.2 | GO:0070458 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.5 | 1.4 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.4 | 3.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.4 | 4.2 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.4 | 8.2 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.4 | 2.1 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.4 | 3.3 | GO:0015074 | DNA integration(GO:0015074) |
| 0.3 | 1.7 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.3 | 1.0 | GO:1903225 | regulation of endodermal cell fate specification(GO:0042663) negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.3 | 5.8 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.3 | 1.9 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.3 | 1.6 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.3 | 6.1 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.3 | 0.5 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 0.3 | 2.5 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.2 | 1.5 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.2 | 3.2 | GO:0042711 | maternal behavior(GO:0042711) |
| 0.2 | 0.7 | GO:0044340 | canonical Wnt signaling pathway involved in regulation of cell proliferation(GO:0044340) |
| 0.2 | 5.7 | GO:0071357 | type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.2 | 2.6 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.2 | 0.7 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.2 | 0.6 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.2 | 2.9 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.2 | 1.8 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) negative regulation of protein folding(GO:1903333) |
| 0.2 | 0.9 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.2 | 0.7 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.2 | 1.0 | GO:0071455 | cellular response to increased oxygen levels(GO:0036295) cellular response to hyperoxia(GO:0071455) |
| 0.2 | 0.9 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.2 | 2.4 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.2 | 0.5 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.2 | 2.8 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 0.4 | GO:0002925 | positive regulation of chronic inflammatory response(GO:0002678) positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) negative regulation of alkaline phosphatase activity(GO:0010693) regulation of branching involved in lung morphogenesis(GO:0061046) necroptotic signaling pathway(GO:0097527) |
| 0.1 | 1.3 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 4.4 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 0.7 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 0.7 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) enamel mineralization(GO:0070166) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 3.8 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.1 | 3.1 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.1 | 3.9 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.1 | 3.0 | GO:0042474 | middle ear morphogenesis(GO:0042474) |
| 0.1 | 9.7 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.1 | 1.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 2.2 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.1 | 0.9 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.1 | 2.9 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 1.7 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 0.3 | GO:0060729 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 3.4 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.1 | 5.0 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.1 | 0.6 | GO:0036506 | maintenance of unfolded protein(GO:0036506) protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.1 | 0.4 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 2.3 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.1 | 0.5 | GO:0031424 | keratinization(GO:0031424) establishment of skin barrier(GO:0061436) |
| 0.1 | 0.4 | GO:0000467 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.1 | 2.7 | GO:0007628 | adult walking behavior(GO:0007628) walking behavior(GO:0090659) |
| 0.1 | 1.2 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 3.7 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.0 | 1.5 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.6 | GO:0010388 | cullin deneddylation(GO:0010388) |
| 0.0 | 0.6 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 1.5 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 1.8 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 2.0 | GO:0002209 | behavioral fear response(GO:0001662) behavioral defense response(GO:0002209) |
| 0.0 | 0.7 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 5.9 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 1.2 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 0.5 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.1 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 2.1 | GO:0000725 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.0 | 1.2 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.4 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 0.7 | GO:0035058 | nonmotile primary cilium assembly(GO:0035058) |
| 0.0 | 0.7 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0045213 | neurotransmitter receptor metabolic process(GO:0045213) |
| 0.0 | 0.6 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.1 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 3.3 | GO:0000398 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.0 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.7 | 9.6 | GO:0043203 | axon hillock(GO:0043203) |
| 0.6 | 3.9 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.6 | 9.0 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.5 | 4.8 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.5 | 11.3 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.4 | 1.9 | GO:0005861 | troponin complex(GO:0005861) |
| 0.4 | 3.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.3 | 2.4 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.3 | 4.0 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.3 | 4.5 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.3 | 2.0 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.2 | 3.5 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.2 | 0.7 | GO:1990879 | CST complex(GO:1990879) |
| 0.2 | 1.0 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.2 | 2.4 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.2 | 0.5 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.2 | 1.2 | GO:0044613 | nuclear pore central transport channel(GO:0044613) Flemming body(GO:0090543) |
| 0.1 | 4.0 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 1.7 | GO:0048188 | MLL3/4 complex(GO:0044666) Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 0.7 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 2.9 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 6.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.9 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 1.7 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 1.8 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.7 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.4 | GO:1990462 | omegasome(GO:1990462) |
| 0.1 | 0.6 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 0.4 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 2.6 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 1.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 1.1 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 2.1 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 1.5 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 3.2 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 18.0 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 2.5 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 1.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.0 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 3.9 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 1.9 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 1.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 2.2 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.5 | GO:0098803 | respiratory chain complex(GO:0098803) |
| 0.0 | 0.7 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.9 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.3 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 15.4 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.6 | GO:0005770 | late endosome(GO:0005770) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.0 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 1.5 | 9.0 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 1.2 | 13.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 1.2 | 3.6 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 1.1 | 3.2 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 1.0 | 4.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.6 | 1.9 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.6 | 3.8 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.6 | 1.8 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.6 | 11.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.5 | 8.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.5 | 2.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.4 | 7.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.4 | 3.8 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.4 | 1.5 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.4 | 1.5 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.4 | 2.2 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.3 | 6.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.3 | 7.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.3 | 2.5 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.3 | 3.1 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.3 | 1.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.2 | 4.6 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.2 | 2.4 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.2 | 3.5 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.2 | 0.7 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.2 | 9.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 2.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.2 | 1.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.2 | 4.8 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.2 | 4.5 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 0.5 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) oncostatin-M receptor activity(GO:0004924) |
| 0.2 | 2.0 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.2 | 4.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 2.9 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 3.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.8 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 6.3 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.1 | 3.2 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.1 | 3.0 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 0.9 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.1 | 6.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 1.0 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 0.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.6 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 1.7 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 20.6 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.1 | 0.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.3 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 0.9 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 0.7 | GO:0070181 | mRNA 5'-UTR binding(GO:0048027) small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 1.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.5 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 0.7 | GO:0010521 | telomerase inhibitor activity(GO:0010521) single-stranded telomeric DNA binding(GO:0043047) |
| 0.1 | 0.7 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 0.4 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.7 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 1.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 3.0 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 1.6 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 3.7 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 3.9 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 2.2 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 4.5 | GO:0000982 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) |
| 0.0 | 1.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 2.4 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 0.9 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 2.4 | GO:0044212 | transcription regulatory region DNA binding(GO:0044212) |
| 0.0 | 1.4 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 1.7 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 5.9 | GO:0016791 | phosphatase activity(GO:0016791) |
| 0.0 | 1.0 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.8 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 9.7 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 0.5 | GO:0034945 | dihydrolipoamide branched chain acyltransferase activity(GO:0004147) palmitoleoyl [acyl-carrier-protein]-dependent acyltransferase activity(GO:0008951) serine O-acyltransferase activity(GO:0016412) O-succinyltransferase activity(GO:0016750) sinapoyltransferase activity(GO:0016752) O-sinapoyltransferase activity(GO:0016753) peptidyl-lysine N6-myristoyltransferase activity(GO:0018030) peptidyl-lysine N6-palmitoyltransferase activity(GO:0018031) benzoyl acetate-CoA thiolase activity(GO:0018711) 3-hydroxybutyryl-CoA thiolase activity(GO:0018712) 3-ketopimelyl-CoA thiolase activity(GO:0018713) N-palmitoyltransferase activity(GO:0019105) acyl-CoA N-acyltransferase activity(GO:0019186) protein-cysteine S-myristoyltransferase activity(GO:0019705) glucosaminyl-phosphotidylinositol O-acyltransferase activity(GO:0032216) ergosterol O-acyltransferase activity(GO:0034737) lanosterol O-acyltransferase activity(GO:0034738) naphthyl-2-oxomethyl-succinyl-CoA succinyl transferase activity(GO:0034848) 2,4,4-trimethyl-3-oxopentanoyl-CoA 2-C-propanoyl transferase activity(GO:0034851) 2-methylhexanoyl-CoA C-acetyltransferase activity(GO:0034915) butyryl-CoA 2-C-propionyltransferase activity(GO:0034919) 2,6-dimethyl-5-methylene-3-oxo-heptanoyl-CoA C-acetyltransferase activity(GO:0034945) L-2-aminoadipate N-acetyltransferase activity(GO:0043741) keto acid formate lyase activity(GO:0043806) azetidine-2-carboxylic acid acetyltransferase activity(GO:0046941) peptidyl-lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0052858) acetyl-CoA:L-lysine N6-acetyltransferase(GO:0090595) |
| 0.0 | 0.6 | GO:0003690 | double-stranded DNA binding(GO:0003690) |
| 0.0 | 1.0 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.2 | GO:0050253 | prenylcysteine methylesterase activity(GO:0010296) 1-oxa-2-oxocycloheptane lactonase activity(GO:0018731) sulfolactone hydrolase activity(GO:0018732) 3,4-dihydrocoumarin hydrolase activity(GO:0018733) butyrolactone hydrolase activity(GO:0018734) endosulfan lactone lactonase activity(GO:0034892) L-ascorbate 6-phosphate lactonase activity(GO:0035460) Ser-tRNA(Thr) hydrolase activity(GO:0043905) Ala-tRNA(Pro) hydrolase activity(GO:0043906) Cys-tRNA(Pro) hydrolase activity(GO:0043907) Ser(Gly)-tRNA(Ala) hydrolase activity(GO:0043908) all-trans-retinyl-palmitate hydrolase, all-trans-retinol forming activity(GO:0047376) retinyl-palmitate esterase activity(GO:0050253) mannosyl-oligosaccharide 1,6-alpha-mannosidase activity(GO:0052767) mannosyl-oligosaccharide 1,3-alpha-mannosidase activity(GO:0052768) methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.0 | 0.4 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 2.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |