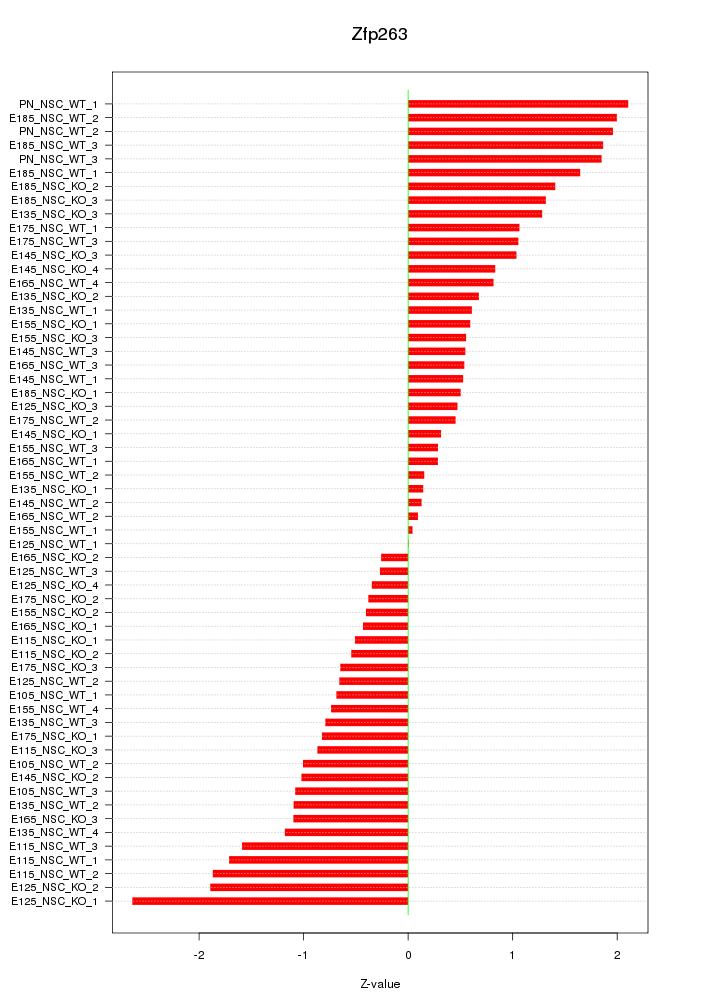
Motif ID: Zfp263
Z-value: 1.066

Transcription factors associated with Zfp263:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Zfp263 | ENSMUSG00000022529.5 | Zfp263 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp263 | mm10_v2_chr16_+_3744089_3744145 | 0.54 | 1.2e-05 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.0 | 18.1 | GO:0072223 | metanephric glomerular mesangium development(GO:0072223) metanephric glomerular mesangial cell proliferation involved in metanephros development(GO:0072262) |
| 3.0 | 9.0 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 2.3 | 11.7 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 1.8 | 8.9 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 1.6 | 4.9 | GO:0043465 | regulation of fermentation(GO:0043465) negative regulation of fermentation(GO:1901003) |
| 1.6 | 6.4 | GO:0006867 | asparagine transport(GO:0006867) glutamine transport(GO:0006868) |
| 1.4 | 10.0 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 1.4 | 5.7 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 1.3 | 5.2 | GO:0099624 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) atrial cardiac muscle cell membrane repolarization(GO:0099624) |
| 1.2 | 3.7 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) ceramide transport(GO:0035627) |
| 1.1 | 3.4 | GO:0043379 | memory T cell differentiation(GO:0043379) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 1.1 | 3.3 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 1.1 | 7.7 | GO:0034047 | regulation of protein phosphatase type 2A activity(GO:0034047) |
| 1.1 | 5.5 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 1.1 | 3.2 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 1.0 | 3.1 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.9 | 3.6 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.9 | 2.7 | GO:0035622 | intrahepatic bile duct development(GO:0035622) renal vesicle induction(GO:0072034) |
| 0.9 | 3.5 | GO:0046469 | platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.9 | 2.6 | GO:0050748 | negative regulation of receptor biosynthetic process(GO:0010871) negative regulation of lipoprotein metabolic process(GO:0050748) |
| 0.9 | 2.6 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.9 | 2.6 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.8 | 16.8 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.8 | 4.9 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.8 | 2.4 | GO:0061535 | glutamate secretion, neurotransmission(GO:0061535) |
| 0.8 | 4.6 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.7 | 2.2 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.7 | 3.6 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.7 | 1.4 | GO:0048715 | negative regulation of oligodendrocyte differentiation(GO:0048715) |
| 0.7 | 4.2 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.7 | 2.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.7 | 4.8 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.7 | 4.1 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.7 | 2.6 | GO:0021564 | glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 0.6 | 3.8 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.6 | 1.9 | GO:1905050 | positive regulation of metalloendopeptidase activity(GO:1904685) positive regulation of metallopeptidase activity(GO:1905050) |
| 0.6 | 6.0 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.6 | 2.4 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.6 | 1.8 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.6 | 1.8 | GO:0035864 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.6 | 1.8 | GO:0072076 | hyaluranon cable assembly(GO:0036118) nephrogenic mesenchyme development(GO:0072076) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.6 | 3.5 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.6 | 1.7 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.6 | 4.0 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.5 | 2.7 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.5 | 1.6 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.5 | 2.1 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.5 | 2.1 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.5 | 3.7 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.5 | 1.5 | GO:0030827 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.5 | 2.0 | GO:0035624 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.5 | 3.0 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.5 | 4.8 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.5 | 5.9 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.4 | 6.5 | GO:0060746 | parental behavior(GO:0060746) |
| 0.4 | 1.3 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.4 | 1.2 | GO:0021893 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 0.4 | 2.1 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.4 | 4.1 | GO:0051608 | organic cation transport(GO:0015695) histamine transport(GO:0051608) |
| 0.4 | 18.1 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.4 | 1.6 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.4 | 0.7 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.4 | 1.4 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.3 | 1.0 | GO:0090298 | positive regulation of fat cell proliferation(GO:0070346) negative regulation of mitochondrial DNA replication(GO:0090298) |
| 0.3 | 2.1 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.3 | 1.0 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.3 | 2.9 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.3 | 3.3 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.3 | 1.0 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.3 | 1.6 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.3 | 6.5 | GO:0006415 | translational termination(GO:0006415) |
| 0.3 | 2.2 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.3 | 3.7 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.3 | 4.8 | GO:0048679 | regulation of axon regeneration(GO:0048679) |
| 0.3 | 1.2 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.3 | 2.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.3 | 2.7 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.3 | 0.3 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.3 | 1.4 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.3 | 3.8 | GO:0042094 | interleukin-2 biosynthetic process(GO:0042094) |
| 0.3 | 4.3 | GO:0042428 | serotonin metabolic process(GO:0042428) |
| 0.3 | 1.6 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.3 | 4.5 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.3 | 0.8 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.3 | 0.5 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.3 | 2.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.2 | 6.2 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.2 | 1.0 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.2 | 1.2 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.2 | 1.7 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.2 | 1.9 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.2 | 1.6 | GO:0051036 | regulation of endosome size(GO:0051036) receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.2 | 0.7 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.2 | 1.1 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.2 | 0.6 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.2 | 3.6 | GO:0034142 | toll-like receptor 4 signaling pathway(GO:0034142) |
| 0.2 | 1.0 | GO:0032911 | nerve growth factor production(GO:0032902) negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.2 | 3.6 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.2 | 1.8 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.2 | 1.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 1.1 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.2 | 1.1 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.2 | 0.5 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.2 | 2.4 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.2 | 1.3 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.2 | 1.2 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.2 | 0.7 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.2 | 0.8 | GO:0002182 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.2 | 2.0 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.2 | 1.3 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.2 | 7.2 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.2 | 1.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.2 | 0.6 | GO:0090148 | membrane fission(GO:0090148) |
| 0.2 | 0.3 | GO:0002069 | columnar/cuboidal epithelial cell maturation(GO:0002069) |
| 0.2 | 0.5 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.2 | 0.6 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 | 1.3 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 0.3 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.1 | 0.4 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.1 | 0.7 | GO:0033136 | serine phosphorylation of STAT3 protein(GO:0033136) |
| 0.1 | 1.0 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 | 0.8 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.1 | 2.0 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.1 | 1.6 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.1 | 1.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.9 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 | 0.9 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.1 | 0.5 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.1 | 0.7 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 | 3.4 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.1 | 0.5 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 | 0.7 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.1 | 0.2 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.1 | 0.8 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 0.7 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 1.1 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 6.5 | GO:0008542 | visual learning(GO:0008542) |
| 0.1 | 0.7 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.1 | 2.4 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 4.0 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.1 | 1.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 8.7 | GO:0051592 | response to calcium ion(GO:0051592) |
| 0.1 | 3.7 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 0.5 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 0.8 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 2.3 | GO:1901798 | positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.1 | 0.7 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 3.9 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.1 | 1.0 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 0.8 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 0.8 | GO:0042473 | outer ear morphogenesis(GO:0042473) |
| 0.1 | 0.2 | GO:0061009 | common bile duct development(GO:0061009) |
| 0.1 | 2.2 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.1 | 0.5 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.7 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 0.2 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.1 | 0.4 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 0.4 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 0.4 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 0.9 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.1 | 0.3 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.1 | 0.3 | GO:2000325 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.1 | 0.5 | GO:0099515 | actin filament-based transport(GO:0099515) |
| 0.1 | 0.9 | GO:0042297 | vocal learning(GO:0042297) imitative learning(GO:0098596) learned vocalization behavior or vocal learning(GO:0098598) |
| 0.1 | 2.7 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.1 | 3.3 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 1.7 | GO:0071242 | cellular response to ammonium ion(GO:0071242) |
| 0.1 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 1.5 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 3.8 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.1 | 1.1 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 0.9 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 3.7 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.6 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.6 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.5 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.3 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 1.0 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.0 | 0.3 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) regulation of spindle organization(GO:0090224) |
| 0.0 | 0.9 | GO:0050679 | positive regulation of epithelial cell proliferation(GO:0050679) |
| 0.0 | 2.2 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.1 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.1 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 1.3 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.2 | GO:2000258 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 0.2 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.0 | 2.2 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.2 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 1.7 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.4 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.3 | GO:0030828 | receptor guanylyl cyclase signaling pathway(GO:0007168) positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.0 | 0.2 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.1 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 1.2 | GO:0042594 | response to starvation(GO:0042594) |
| 0.0 | 1.6 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.3 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 1.0 | GO:0014911 | positive regulation of smooth muscle cell migration(GO:0014911) |
| 0.0 | 1.4 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.2 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.5 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.1 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.0 | 0.7 | GO:0060043 | regulation of cardiac muscle cell proliferation(GO:0060043) |
| 0.0 | 3.9 | GO:0006813 | potassium ion transport(GO:0006813) |
| 0.0 | 0.6 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.1 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.0 | 0.6 | GO:2000058 | regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000058) |
| 0.0 | 0.3 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.0 | 0.1 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.0 | 0.8 | GO:0071346 | cellular response to interferon-gamma(GO:0071346) |
| 0.0 | 0.7 | GO:0048008 | platelet-derived growth factor receptor signaling pathway(GO:0048008) |
| 0.0 | 0.1 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 1.0 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 0.0 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.0 | 0.4 | GO:0031114 | negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.3 | GO:0006956 | complement activation(GO:0006956) |
| 0.0 | 0.1 | GO:0060693 | regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.0 | 1.0 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.1 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 7.0 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 1.0 | 3.9 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.9 | 2.6 | GO:0034684 | integrin alphav-beta5 complex(GO:0034684) |
| 0.7 | 4.0 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.7 | 10.0 | GO:0043196 | varicosity(GO:0043196) |
| 0.6 | 6.1 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.5 | 1.0 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.5 | 7.0 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.4 | 4.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.4 | 2.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.4 | 2.1 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.4 | 2.1 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.4 | 7.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.4 | 4.0 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.4 | 2.6 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.3 | 1.7 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.3 | 0.9 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.3 | 5.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.3 | 1.4 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.3 | 0.8 | GO:0031417 | NatC complex(GO:0031417) |
| 0.3 | 3.8 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.3 | 1.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.2 | 1.5 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.2 | 1.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 0.4 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.2 | 13.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.2 | 2.2 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.2 | 1.6 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.2 | 0.5 | GO:1990075 | kinesin II complex(GO:0016939) periciliary membrane compartment(GO:1990075) |
| 0.2 | 12.1 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.2 | 0.8 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 0.8 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 1.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 2.6 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 3.7 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 1.7 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 4.1 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 2.2 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 4.3 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 5.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 0.8 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 0.3 | GO:0044299 | C-fiber(GO:0044299) |
| 0.1 | 1.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 1.1 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 1.1 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 0.3 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.1 | 0.7 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.1 | 4.3 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 1.0 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 0.7 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 1.5 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.7 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 0.8 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 5.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 0.7 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 6.9 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 7.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 4.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 0.5 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 1.1 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 0.9 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 0.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 1.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.9 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 10.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 4.0 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.7 | GO:0044217 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 3.4 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 3.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.2 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 2.0 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 8.9 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.5 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.6 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 1.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 5.5 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.3 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.2 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 28.1 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 0.3 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.5 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 1.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.0 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.5 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 2.2 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.1 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.9 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 2.1 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.9 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 0.3 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.5 | GO:0031252 | cell leading edge(GO:0031252) |
| 0.0 | 1.3 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.7 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 6.6 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 1.0 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 4.7 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.3 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.8 | GO:0005814 | centriole(GO:0005814) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 16.0 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 2.6 | 15.6 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 2.1 | 6.4 | GO:0043912 | D-lysine oxidase activity(GO:0043912) |
| 1.6 | 6.5 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 1.5 | 5.9 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 1.4 | 4.1 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 1.3 | 6.4 | GO:0015186 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 1.2 | 4.9 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 1.2 | 3.5 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 1.1 | 6.8 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 1.1 | 8.8 | GO:0103116 | alpha-D-galactofuranose transporter activity(GO:0103116) |
| 1.0 | 5.2 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 1.0 | 8.9 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.9 | 11.3 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.9 | 15.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.9 | 2.6 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.8 | 10.0 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.8 | 2.3 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.7 | 3.7 | GO:0097001 | sphingolipid transporter activity(GO:0046624) ceramide binding(GO:0097001) |
| 0.7 | 2.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.7 | 3.5 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.7 | 2.0 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) epinephrine binding(GO:0051379) |
| 0.7 | 3.3 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.6 | 6.0 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.6 | 8.4 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.6 | 3.0 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.6 | 3.5 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.5 | 1.6 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.5 | 2.6 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) neuregulin binding(GO:0038132) |
| 0.5 | 5.8 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.5 | 3.2 | GO:0009374 | biotin binding(GO:0009374) |
| 0.5 | 4.1 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) potassium ion antiporter activity(GO:0022821) |
| 0.5 | 1.5 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.5 | 3.6 | GO:0031432 | titin binding(GO:0031432) |
| 0.4 | 2.9 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.4 | 1.2 | GO:0036361 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.4 | 1.2 | GO:0004008 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.4 | 3.5 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.4 | 1.6 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.4 | 2.3 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.4 | 2.7 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.4 | 3.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.4 | 1.8 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.4 | 1.4 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.4 | 3.9 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.4 | 1.8 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.3 | 3.7 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.3 | 1.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.3 | 4.4 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.3 | 3.4 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.3 | 1.2 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.3 | 0.9 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) RNA strand annealing activity(GO:0033592) |
| 0.3 | 3.8 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.3 | 1.2 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.3 | 1.7 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.3 | 4.0 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.2 | 1.0 | GO:0071532 | ornithine decarboxylase inhibitor activity(GO:0008073) ankyrin repeat binding(GO:0071532) |
| 0.2 | 1.4 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.2 | 0.9 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.2 | 0.7 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.2 | 0.9 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.2 | 2.8 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.2 | 1.3 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 3.6 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.2 | 1.9 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.2 | 0.8 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.2 | 2.0 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.2 | 7.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 1.9 | GO:0042805 | actinin binding(GO:0042805) |
| 0.2 | 0.7 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.2 | 3.8 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.2 | 1.0 | GO:0015375 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) glycine:sodium symporter activity(GO:0015375) |
| 0.2 | 1.6 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
| 0.2 | 1.7 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.2 | 0.6 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 0.7 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 4.2 | GO:0022824 | transmitter-gated ion channel activity(GO:0022824) transmitter-gated channel activity(GO:0022835) |
| 0.1 | 5.7 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 1.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 1.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 3.0 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 1.0 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 2.6 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 1.4 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 2.1 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 1.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 1.7 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.1 | 0.5 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.4 | GO:0001639 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) |
| 0.1 | 0.6 | GO:0050543 | icosatetraenoic acid binding(GO:0050543) arachidonic acid binding(GO:0050544) |
| 0.1 | 0.8 | GO:0016215 | acyl-CoA desaturase activity(GO:0016215) |
| 0.1 | 2.0 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 1.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 1.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 2.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.5 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 0.8 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.8 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 3.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 1.1 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 2.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 0.4 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 2.7 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.1 | 0.3 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 0.3 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 2.0 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 1.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 1.0 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 0.2 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 1.9 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.3 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 1.2 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 1.3 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.1 | 0.7 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 0.7 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 1.2 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.9 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.9 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 3.4 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 0.2 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) |
| 0.0 | 0.3 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.5 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.3 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.1 | GO:0004368 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) |
| 0.0 | 0.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.7 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.3 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 1.3 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.7 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 1.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 2.0 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.1 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.0 | 0.6 | GO:0015645 | fatty acid ligase activity(GO:0015645) |
| 0.0 | 0.8 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.3 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 1.3 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 1.3 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.3 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 1.0 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.6 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.9 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.5 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.4 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 1.5 | GO:0101005 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.2 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.0 | 0.1 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 1.4 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.5 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.6 | GO:0019003 | GDP binding(GO:0019003) |