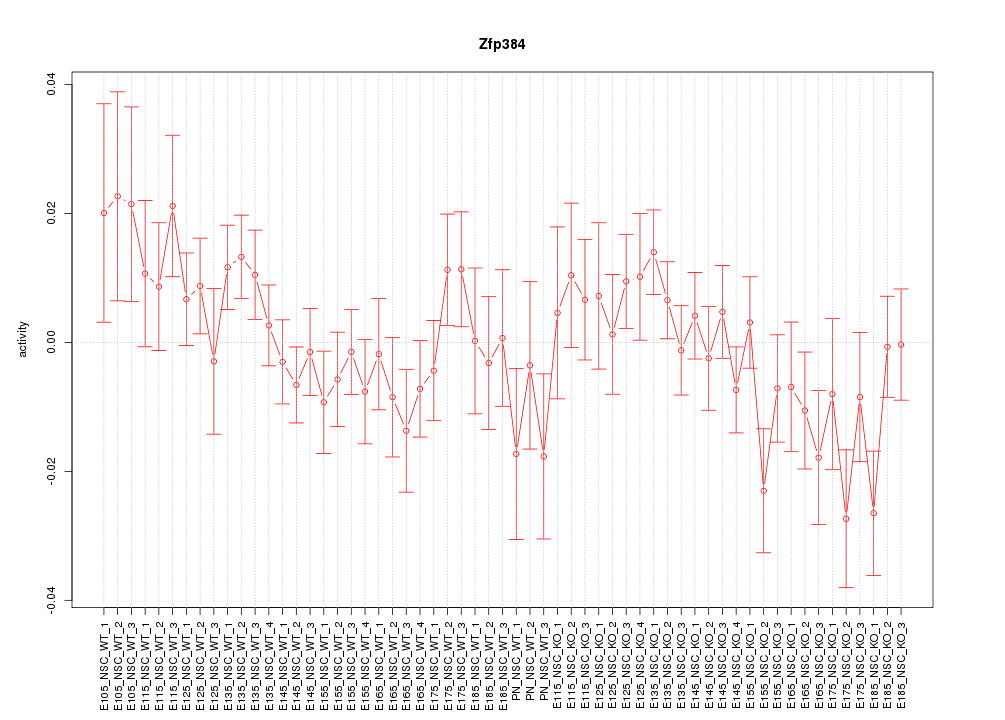
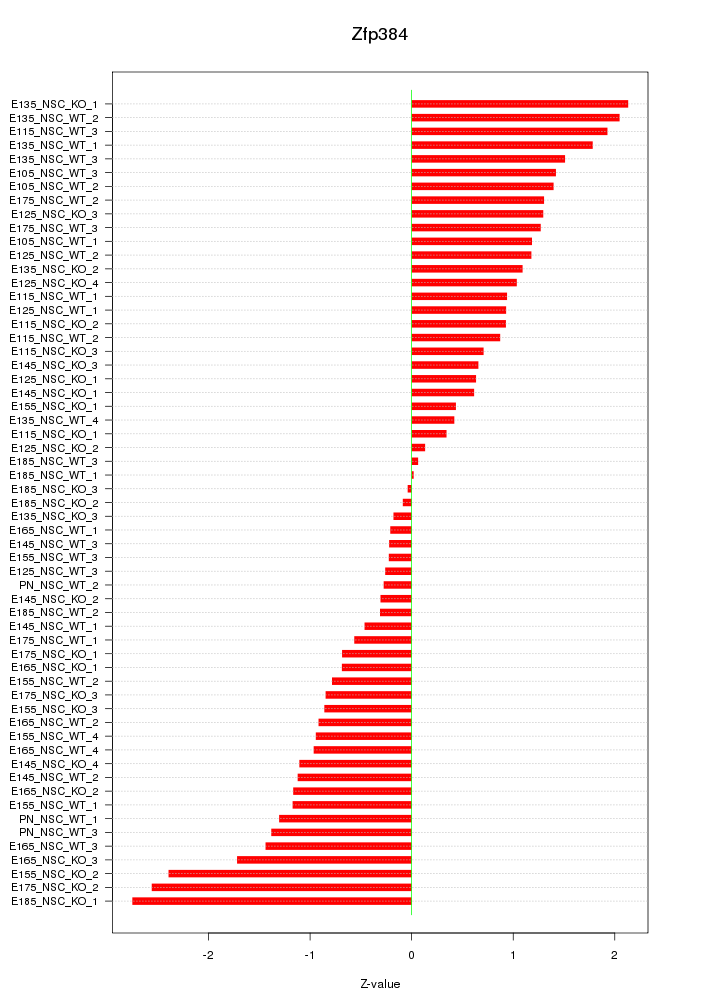
Motif ID: Zfp384
Z-value: 1.149
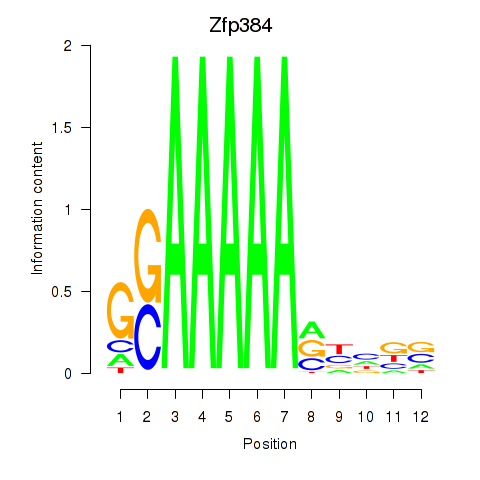
Transcription factors associated with Zfp384:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Zfp384 | ENSMUSG00000038346.12 | Zfp384 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp384 | mm10_v2_chr6_+_125009261_125009317 | -0.23 | 8.4e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 25.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 2.5 | 7.5 | GO:0097402 | neuroblast migration(GO:0097402) |
| 2.1 | 6.2 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 2.0 | 2.0 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 1.7 | 5.1 | GO:0019405 | alditol catabolic process(GO:0019405) |
| 1.6 | 7.9 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 1.4 | 4.3 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 1.3 | 5.0 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 1.2 | 3.6 | GO:0034334 | adherens junction maintenance(GO:0034334) intermediate filament bundle assembly(GO:0045110) maintenance of organ identity(GO:0048496) |
| 1.1 | 2.2 | GO:0046370 | fructose biosynthetic process(GO:0046370) |
| 1.0 | 3.1 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 1.0 | 3.1 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 1.0 | 2.0 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 1.0 | 9.8 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 1.0 | 3.8 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.9 | 2.8 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 0.9 | 2.8 | GO:0016095 | polyprenol catabolic process(GO:0016095) terpenoid catabolic process(GO:0016115) primary alcohol catabolic process(GO:0034310) |
| 0.9 | 2.7 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.9 | 2.7 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.8 | 2.5 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.8 | 2.5 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.8 | 3.3 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.8 | 2.4 | GO:0003104 | positive regulation of glomerular filtration(GO:0003104) cell communication by chemical coupling(GO:0010643) |
| 0.8 | 3.0 | GO:0035795 | negative regulation of mitochondrial membrane permeability(GO:0035795) |
| 0.7 | 5.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.7 | 7.2 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.7 | 2.2 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.7 | 2.1 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.7 | 1.4 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) |
| 0.7 | 1.3 | GO:0032066 | nucleolus to nucleoplasm transport(GO:0032066) |
| 0.7 | 0.7 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.6 | 9.1 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.6 | 1.9 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.6 | 1.8 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.6 | 1.8 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.6 | 3.0 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.6 | 4.8 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.6 | 6.5 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.6 | 5.8 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.6 | 1.7 | GO:0040009 | regulation of growth rate(GO:0040009) protein poly-ADP-ribosylation(GO:0070212) |
| 0.6 | 2.8 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.6 | 3.4 | GO:0002553 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.5 | 2.2 | GO:0046084 | adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.5 | 1.6 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.5 | 1.1 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.5 | 1.6 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.5 | 10.4 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.5 | 2.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.5 | 1.0 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.5 | 1.5 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.5 | 2.0 | GO:1903898 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.5 | 6.1 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.5 | 1.5 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.5 | 1.5 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.5 | 0.5 | GO:0033122 | regulation of purine nucleotide catabolic process(GO:0033121) negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 0.5 | 1.5 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.5 | 3.3 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.5 | 6.8 | GO:0009191 | ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.5 | 2.3 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.4 | 2.2 | GO:0019659 | lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.4 | 3.1 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.4 | 1.3 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.4 | 1.3 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.4 | 1.7 | GO:0045343 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 0.4 | 1.3 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.4 | 1.3 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.4 | 2.1 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.4 | 2.9 | GO:0051461 | regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.4 | 5.0 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 0.4 | 1.2 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.4 | 2.5 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.4 | 1.6 | GO:0046210 | nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.4 | 1.2 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.4 | 2.0 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.4 | 0.4 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.4 | 2.0 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.4 | 4.0 | GO:0046036 | GTP biosynthetic process(GO:0006183) CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.4 | 1.6 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.4 | 2.0 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.4 | 1.2 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.4 | 1.9 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.4 | 0.8 | GO:1990705 | cholangiocyte proliferation(GO:1990705) |
| 0.4 | 1.5 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.4 | 1.2 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.4 | 1.5 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.4 | 1.9 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.4 | 1.1 | GO:0006296 | base-excision repair, AP site formation(GO:0006285) nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.4 | 1.8 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.4 | 1.1 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.4 | 4.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.4 | 1.1 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.3 | 3.1 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.3 | 1.4 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.3 | 1.0 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.3 | 1.7 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.3 | 2.0 | GO:0060290 | transdifferentiation(GO:0060290) |
| 0.3 | 2.0 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.3 | 4.2 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.3 | 2.2 | GO:0070458 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.3 | 1.0 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.3 | 1.3 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.3 | 2.2 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.3 | 1.2 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.3 | 0.9 | GO:0050929 | corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.3 | 1.8 | GO:1902965 | protein localization to early endosome(GO:1902946) regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.3 | 0.9 | GO:1901228 | positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.3 | 0.9 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.3 | 1.8 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.3 | 2.4 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.3 | 1.2 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.3 | 0.9 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.3 | 0.9 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.3 | 1.5 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.3 | 2.3 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.3 | 1.7 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.3 | 0.6 | GO:0014735 | regulation of muscle atrophy(GO:0014735) |
| 0.3 | 1.7 | GO:1903755 | regulation of SUMO transferase activity(GO:1903182) positive regulation of SUMO transferase activity(GO:1903755) |
| 0.3 | 0.8 | GO:0045014 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.3 | 8.6 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.3 | 1.4 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.3 | 0.8 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.3 | 0.8 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.3 | 2.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.3 | 5.9 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.3 | 1.6 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.3 | 2.4 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.3 | 2.4 | GO:0085020 | protein K6-linked ubiquitination(GO:0085020) |
| 0.3 | 2.1 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.3 | 0.8 | GO:2000832 | cortisol secretion(GO:0043400) regulation of cortisol secretion(GO:0051462) negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.3 | 0.8 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.3 | 0.3 | GO:0006404 | RNA import into nucleus(GO:0006404) |
| 0.3 | 1.6 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.3 | 0.8 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.3 | 1.0 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.3 | 0.3 | GO:0014891 | striated muscle atrophy(GO:0014891) |
| 0.2 | 0.7 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.2 | 1.6 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.2 | 1.2 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.2 | 3.3 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.2 | 0.7 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.2 | 2.3 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.2 | 2.7 | GO:0021794 | thalamus development(GO:0021794) |
| 0.2 | 0.9 | GO:0045915 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.2 | 2.7 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.2 | 1.8 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.2 | 0.2 | GO:0002586 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.2 | 1.6 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.2 | 0.7 | GO:1900149 | positive regulation of Schwann cell migration(GO:1900149) |
| 0.2 | 1.3 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.2 | 0.9 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.2 | 0.6 | GO:0061743 | motor learning(GO:0061743) |
| 0.2 | 0.9 | GO:0033580 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.2 | 0.6 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.2 | 2.8 | GO:0071281 | cellular response to iron ion(GO:0071281) |
| 0.2 | 0.9 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.2 | 0.6 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.2 | 4.2 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.2 | 1.0 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.2 | 0.6 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.2 | 1.0 | GO:0051096 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.2 | 1.7 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.2 | 3.1 | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.2 | 2.9 | GO:1903672 | positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.2 | 0.6 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.2 | 0.8 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.2 | 0.4 | GO:0002678 | positive regulation of chronic inflammatory response(GO:0002678) |
| 0.2 | 1.8 | GO:2000269 | regulation of fibroblast apoptotic process(GO:2000269) |
| 0.2 | 2.9 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.2 | 1.4 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.2 | 0.2 | GO:1900110 | negative regulation of histone H3-K9 dimethylation(GO:1900110) |
| 0.2 | 1.0 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.2 | 1.0 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.2 | 1.7 | GO:1901970 | positive regulation of mitotic sister chromatid separation(GO:1901970) |
| 0.2 | 1.3 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.2 | 1.3 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.2 | 0.9 | GO:0031424 | keratinization(GO:0031424) |
| 0.2 | 2.2 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.2 | 0.5 | GO:0031990 | mRNA export from nucleus in response to heat stress(GO:0031990) |
| 0.2 | 0.5 | GO:0050904 | diapedesis(GO:0050904) |
| 0.2 | 3.4 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.2 | 1.3 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.2 | 3.6 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.2 | 0.5 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.2 | 0.4 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.2 | 0.3 | GO:0060023 | soft palate development(GO:0060023) |
| 0.2 | 4.2 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.2 | 1.0 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.2 | 0.7 | GO:1904008 | response to monosodium glutamate(GO:1904008) cellular response to monosodium glutamate(GO:1904009) |
| 0.2 | 1.4 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.2 | 0.3 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.2 | 0.9 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.2 | 1.0 | GO:0015684 | ferrous iron transport(GO:0015684) |
| 0.2 | 2.5 | GO:0007530 | sex determination(GO:0007530) |
| 0.2 | 0.7 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.2 | 1.2 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.2 | 0.7 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.2 | 2.0 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.2 | 0.7 | GO:0006547 | histidine metabolic process(GO:0006547) |
| 0.2 | 4.9 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.2 | 1.3 | GO:0036506 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.2 | 0.3 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.2 | 0.5 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.2 | 0.8 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.2 | 1.1 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.2 | 2.4 | GO:0007063 | regulation of sister chromatid cohesion(GO:0007063) |
| 0.2 | 1.4 | GO:0007512 | adult heart development(GO:0007512) |
| 0.2 | 0.9 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.2 | 0.9 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.2 | 1.1 | GO:0007320 | insemination(GO:0007320) |
| 0.1 | 0.3 | GO:0002677 | regulation of chronic inflammatory response(GO:0002676) negative regulation of chronic inflammatory response(GO:0002677) |
| 0.1 | 3.6 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.1 | 3.4 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 0.6 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.1 | 0.4 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 | 0.6 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 0.1 | 1.5 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.1 | 1.0 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.1 | 0.9 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.9 | GO:0046909 | intermembrane transport(GO:0046909) protein localization to ciliary membrane(GO:1903441) |
| 0.1 | 0.4 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.1 | 0.4 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 1.4 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 0.4 | GO:0014028 | notochord formation(GO:0014028) |
| 0.1 | 0.3 | GO:0033092 | positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.1 | 0.6 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.1 | 0.7 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.6 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.1 | 0.8 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 0.7 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 1.9 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 0.8 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.1 | 0.5 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.1 | 8.1 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.1 | 1.6 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.1 | 1.6 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.1 | 1.2 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.1 | 1.8 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.1 | 4.6 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 0.4 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 1.4 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.1 | 0.6 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.1 | 1.4 | GO:0000059 | protein import into nucleus, docking(GO:0000059) |
| 0.1 | 0.9 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.1 | 4.4 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.1 | 6.7 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 1.0 | GO:0071426 | ribonucleoprotein complex export from nucleus(GO:0071426) |
| 0.1 | 0.6 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 5.6 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.1 | 0.7 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.1 | 2.1 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.1 | 1.1 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 0.7 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 1.6 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 0.4 | GO:0003273 | cell migration involved in endocardial cushion formation(GO:0003273) |
| 0.1 | 0.7 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 0.1 | 1.7 | GO:0010388 | cullin deneddylation(GO:0010388) |
| 0.1 | 2.6 | GO:0010883 | regulation of lipid storage(GO:0010883) |
| 0.1 | 0.8 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 0.5 | GO:1902047 | polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 0.3 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.1 | 0.5 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.1 | 0.3 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.1 | 2.5 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.1 | 0.2 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.1 | 0.8 | GO:0006273 | lagging strand elongation(GO:0006273) |
| 0.1 | 2.2 | GO:0045840 | positive regulation of mitotic nuclear division(GO:0045840) |
| 0.1 | 1.0 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 0.7 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 3.5 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.1 | 0.8 | GO:2000380 | regulation of mesoderm development(GO:2000380) negative regulation of mesoderm development(GO:2000381) |
| 0.1 | 0.5 | GO:1901409 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 1.2 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.1 | 1.1 | GO:0021924 | cell proliferation in external granule layer(GO:0021924) cerebellar granule cell precursor proliferation(GO:0021930) |
| 0.1 | 1.5 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 1.0 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 0.4 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.1 | 0.2 | GO:0009838 | abscission(GO:0009838) |
| 0.1 | 1.2 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 0.5 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.1 | 0.4 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.1 | 0.1 | GO:0010813 | neuropeptide catabolic process(GO:0010813) |
| 0.1 | 0.4 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.8 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 1.6 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 1.0 | GO:0031999 | negative regulation of fatty acid beta-oxidation(GO:0031999) |
| 0.1 | 0.4 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.1 | 0.3 | GO:1903897 | regulation of PERK-mediated unfolded protein response(GO:1903897) |
| 0.1 | 0.3 | GO:1901896 | negative regulation of action potential(GO:0045759) positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.1 | 0.5 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.1 | 3.9 | GO:0035306 | positive regulation of dephosphorylation(GO:0035306) |
| 0.1 | 0.8 | GO:0000479 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.1 | 0.3 | GO:1903912 | negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.1 | 1.0 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 0.4 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 0.4 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 4.5 | GO:0007628 | adult walking behavior(GO:0007628) walking behavior(GO:0090659) |
| 0.1 | 1.0 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 0.7 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 0.2 | GO:0060027 | cardiac right atrium morphogenesis(GO:0003213) convergent extension involved in gastrulation(GO:0060027) |
| 0.1 | 0.7 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.4 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 1.6 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 0.4 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.1 | 0.6 | GO:0046628 | positive regulation of insulin receptor signaling pathway(GO:0046628) |
| 0.1 | 2.0 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 0.2 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.1 | 0.4 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 | 10.0 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.1 | 0.4 | GO:0061009 | common bile duct development(GO:0061009) |
| 0.1 | 2.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.4 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.1 | 2.7 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 0.7 | GO:0071168 | protein localization to chromatin(GO:0071168) |
| 0.1 | 0.9 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 0.8 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 0.2 | GO:0000965 | mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.1 | 0.9 | GO:0000305 | response to oxygen radical(GO:0000305) |
| 0.1 | 0.4 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) response to amino acid starvation(GO:1990928) |
| 0.1 | 0.2 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.1 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 1.7 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.1 | 3.0 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 1.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 2.2 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.1 | 0.1 | GO:0042663 | negative regulation of cell fate specification(GO:0009996) regulation of endodermal cell fate specification(GO:0042663) |
| 0.1 | 3.4 | GO:2000177 | regulation of neural precursor cell proliferation(GO:2000177) |
| 0.1 | 1.4 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 0.7 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 | 0.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.5 | GO:0048617 | embryonic foregut morphogenesis(GO:0048617) |
| 0.1 | 0.7 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.1 | 1.2 | GO:0045765 | regulation of angiogenesis(GO:0045765) |
| 0.1 | 1.8 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 0.7 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.1 | 0.6 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.1 | 0.4 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 1.9 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.1 | 0.3 | GO:0032346 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 0.1 | 0.9 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.1 | 1.8 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 1.2 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.1 | 0.4 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 0.6 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.1 | 0.4 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 0.4 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 6.6 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.1 | 0.9 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.1 | 0.4 | GO:0030220 | platelet formation(GO:0030220) |
| 0.1 | 0.3 | GO:2000794 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000794) |
| 0.1 | 0.1 | GO:0071166 | ribonucleoprotein complex localization(GO:0071166) |
| 0.1 | 0.3 | GO:0042148 | strand invasion(GO:0042148) |
| 0.1 | 1.1 | GO:0030201 | heparan sulfate proteoglycan metabolic process(GO:0030201) |
| 0.1 | 0.9 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.1 | 0.9 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 0.7 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 1.3 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.1 | 0.1 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.1 | 1.6 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.1 | 0.2 | GO:0030578 | PML body organization(GO:0030578) |
| 0.1 | 0.6 | GO:0035058 | nonmotile primary cilium assembly(GO:0035058) |
| 0.1 | 1.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.8 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.7 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.7 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.1 | 0.5 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 0.5 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.1 | 2.2 | GO:0000725 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.1 | 2.0 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.1 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 2.0 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.5 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:0035696 | monocyte extravasation(GO:0035696) regulation of monocyte extravasation(GO:2000437) |
| 0.0 | 2.5 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.6 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.4 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.4 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.0 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.5 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.2 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 0.4 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.3 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.3 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.9 | GO:0018279 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.8 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.3 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.5 | GO:0035646 | endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.0 | 1.7 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 1.3 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.1 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.2 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.2 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.2 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.4 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.2 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 1.9 | GO:0048663 | neuron fate commitment(GO:0048663) |
| 0.0 | 0.6 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 0.1 | GO:0060903 | regulation of meiosis I(GO:0060631) positive regulation of meiosis I(GO:0060903) |
| 0.0 | 0.1 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.0 | 0.2 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.0 | 0.2 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.2 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.3 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.7 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.2 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.0 | 1.3 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.6 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.0 | 0.2 | GO:0036093 | male germ cell proliferation(GO:0002176) germ cell proliferation(GO:0036093) |
| 0.0 | 0.4 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.5 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.5 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 0.2 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.2 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.5 | GO:1903077 | negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 | 0.2 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.0 | 0.5 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 1.6 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.5 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.0 | 0.4 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 4.1 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.2 | GO:1902742 | apoptotic process involved in development(GO:1902742) |
| 0.0 | 0.2 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.0 | 0.6 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.6 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.7 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.2 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.1 | GO:2000561 | CD4-positive, alpha-beta T cell proliferation(GO:0035739) regulation of natural killer cell degranulation(GO:0043321) positive regulation of defense response to bacterium(GO:1900426) negative regulation of interferon-gamma secretion(GO:1902714) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 1.6 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.4 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.7 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 0.0 | 0.0 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.0 | 2.1 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.0 | 0.2 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 | 0.1 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.3 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.8 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.8 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.0 | 0.1 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.0 | 0.3 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.1 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.0 | 0.3 | GO:0006220 | pyrimidine nucleotide metabolic process(GO:0006220) |
| 0.0 | 0.7 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.3 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.2 | GO:0043312 | neutrophil degranulation(GO:0043312) |
| 0.0 | 0.4 | GO:0050680 | negative regulation of epithelial cell proliferation(GO:0050680) |
| 0.0 | 0.6 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.1 | GO:0061756 | leukocyte tethering or rolling(GO:0050901) leukocyte adhesion to vascular endothelial cell(GO:0061756) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 2.5 | GO:0007059 | chromosome segregation(GO:0007059) |
| 0.0 | 0.4 | GO:1901343 | negative regulation of angiogenesis(GO:0016525) negative regulation of vasculature development(GO:1901343) negative regulation of blood vessel morphogenesis(GO:2000181) |
| 0.0 | 0.1 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.5 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.4 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.1 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.0 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.8 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.0 | 1.2 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 0.4 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.1 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.7 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.5 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 0.2 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.1 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.7 | GO:0007548 | sex differentiation(GO:0007548) |
| 0.0 | 0.4 | GO:0043039 | tRNA aminoacylation(GO:0043039) |
| 0.0 | 0.1 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.2 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.1 | GO:0019395 | fatty acid oxidation(GO:0019395) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 24.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 1.3 | 4.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.9 | 2.7 | GO:0071914 | prominosome(GO:0071914) |
| 0.8 | 2.5 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.8 | 2.3 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.7 | 2.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.7 | 2.9 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.7 | 2.8 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.6 | 1.9 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.6 | 2.4 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.6 | 2.4 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.6 | 0.6 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.6 | 1.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.5 | 3.2 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.5 | 1.6 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.5 | 7.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.5 | 1.5 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.5 | 4.5 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.5 | 2.0 | GO:0030677 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.5 | 2.4 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.5 | 4.2 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.5 | 4.1 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.4 | 2.7 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.4 | 1.3 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 0.4 | 1.7 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.4 | 5.7 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.4 | 1.3 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 0.4 | 2.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.4 | 5.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.4 | 1.6 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.4 | 1.9 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.4 | 2.6 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.4 | 2.1 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.3 | 1.0 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.3 | 2.4 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.3 | 1.6 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.3 | 3.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.3 | 1.6 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.3 | 2.2 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.3 | 2.8 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.3 | 2.5 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.3 | 0.9 | GO:0000811 | GINS complex(GO:0000811) |
| 0.3 | 0.6 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.3 | 0.9 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.3 | 1.4 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.3 | 1.1 | GO:0002111 | BRCA2-BRAF35 complex(GO:0002111) |
| 0.3 | 1.9 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.3 | 5.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.3 | 4.8 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.3 | 0.8 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.3 | 5.9 | GO:0001741 | XY body(GO:0001741) |
| 0.2 | 1.2 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.2 | 1.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.2 | 0.9 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.2 | 3.1 | GO:0042581 | specific granule(GO:0042581) |
| 0.2 | 1.2 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.2 | 16.6 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.2 | 1.8 | GO:0051286 | cell tip(GO:0051286) |
| 0.2 | 1.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 2.9 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.2 | 0.7 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.2 | 2.6 | GO:0048188 | MLL3/4 complex(GO:0044666) Set1C/COMPASS complex(GO:0048188) |
| 0.2 | 4.4 | GO:0030894 | replisome(GO:0030894) |
| 0.2 | 2.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.2 | 2.0 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.2 | 1.4 | GO:0005638 | lamin filament(GO:0005638) |
| 0.2 | 3.1 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.2 | 2.3 | GO:0005902 | microvillus(GO:0005902) |
| 0.2 | 3.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.2 | 2.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 2.5 | GO:0070938 | contractile ring(GO:0070938) |
| 0.2 | 2.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.2 | 0.9 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.2 | 0.9 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 0.5 | GO:0005940 | septin ring(GO:0005940) |
| 0.2 | 0.5 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.2 | 0.7 | GO:0000798 | nuclear cohesin complex(GO:0000798) nuclear meiotic cohesin complex(GO:0034991) |
| 0.2 | 1.4 | GO:0032982 | myosin filament(GO:0032982) |
| 0.2 | 2.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 1.7 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.2 | 1.2 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.2 | 1.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.2 | 0.7 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.2 | 0.8 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 0.6 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.2 | 2.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 1.6 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.2 | 1.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.2 | 1.1 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.2 | 3.2 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 1.5 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.1 | 1.0 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.4 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.1 | 0.4 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.1 | 2.0 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 2.4 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 1.0 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 2.0 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 0.8 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 1.7 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.1 | 1.5 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 1.2 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.1 | 1.8 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.9 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.1 | 0.7 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.1 | 5.2 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 0.5 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 0.8 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 0.4 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.1 | 1.4 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 0.1 | 1.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 2.1 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 1.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 1.5 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 1.6 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.1 | 1.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 11.8 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 0.9 | GO:0001740 | Barr body(GO:0001740) |
| 0.1 | 1.5 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 1.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 1.0 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 9.9 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 1.2 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.1 | 1.0 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.1 | 0.6 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 4.7 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 0.9 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 3.0 | GO:0000152 | nuclear ubiquitin ligase complex(GO:0000152) |
| 0.1 | 1.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.7 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 3.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 4.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 1.1 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 0.8 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 2.0 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 3.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 0.6 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.7 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 4.4 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.8 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.4 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 6.8 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 0.2 | GO:1990393 | 3M complex(GO:1990393) |
| 0.1 | 0.2 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.1 | 0.8 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.8 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.1 | 0.4 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 0.4 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 0.4 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.1 | 0.5 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.1 | 0.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 3.5 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 1.7 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 1.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 0.5 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 0.3 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 0.3 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 0.3 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 2.1 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 1.1 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 0.1 | GO:0070449 | elongin complex(GO:0070449) |
| 0.1 | 0.9 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.1 | 0.9 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.1 | GO:0000178 | cytoplasmic exosome (RNase complex)(GO:0000177) exosome (RNase complex)(GO:0000178) |
| 0.1 | 1.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.5 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 1.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 2.5 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 0.9 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 2.2 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 2.6 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 0.5 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 2.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 0.7 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 1.3 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.1 | 1.3 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.1 | 0.3 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 11.7 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 2.4 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 2.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 1.9 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.4 | GO:0030914 | STAGA complex(GO:0030914) SAGA-type complex(GO:0070461) |
| 0.0 | 0.6 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.7 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.7 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.4 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.5 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 1.6 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.1 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.4 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.0 | 0.7 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 1.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.3 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.9 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 1.9 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 2.8 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 2.3 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.2 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 0.5 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.7 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.5 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 1.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.0 | GO:0030684 | preribosome(GO:0030684) |
| 0.0 | 0.9 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.8 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.4 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.2 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.2 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.5 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 1.0 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 0.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 1.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.7 | GO:0030120 | vesicle coat(GO:0030120) |
| 0.0 | 0.6 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 8.4 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.1 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.5 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.8 | GO:0019867 | mitochondrial outer membrane(GO:0005741) outer membrane(GO:0019867) organelle outer membrane(GO:0031968) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.1 | 24.2 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 1.8 | 5.4 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 1.5 | 4.4 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 1.4 | 9.8 | GO:0070061 | fructose binding(GO:0070061) |
| 1.4 | 4.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 1.3 | 7.9 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 1.1 | 3.3 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 1.0 | 4.2 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.9 | 2.8 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.9 | 2.8 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.9 | 2.8 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.9 | 6.8 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.8 | 1.6 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.8 | 6.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.7 | 2.2 | GO:0018738 | S-formylglutathione hydrolase activity(GO:0018738) |
| 0.7 | 0.7 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.7 | 2.7 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.6 | 1.9 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.6 | 1.9 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.6 | 1.8 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.6 | 1.8 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.6 | 3.0 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.6 | 2.3 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.6 | 2.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.5 | 2.1 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.5 | 2.0 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.5 | 1.5 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.5 | 2.9 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.5 | 3.8 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.5 | 2.4 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.5 | 2.8 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.5 | 2.4 | GO:0071253 | connexin binding(GO:0071253) |
| 0.5 | 2.8 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.4 | 1.3 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.4 | 3.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.4 | 4.3 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.4 | 1.7 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.4 | 1.3 | GO:0071862 | protein phosphatase type 1 activator activity(GO:0071862) |
| 0.4 | 1.2 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) U7 snRNA binding(GO:0071209) |
| 0.4 | 1.6 | GO:0016708 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) iron-cytochrome-c reductase activity(GO:0047726) |
| 0.4 | 4.4 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.4 | 2.4 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.4 | 5.9 | GO:0008430 | selenium binding(GO:0008430) |
| 0.4 | 7.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.4 | 2.3 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.4 | 1.1 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.4 | 1.4 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.4 | 0.4 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.4 | 1.1 | GO:0047598 | 7-dehydrocholesterol reductase activity(GO:0047598) |
| 0.4 | 7.7 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.3 | 2.8 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.3 | 3.1 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.3 | 1.0 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.3 | 3.3 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.3 | 1.6 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.3 | 1.9 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.3 | 1.3 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.3 | 4.7 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.3 | 0.9 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.3 | 1.5 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.3 | 0.3 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.3 | 2.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.3 | 1.8 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.3 | 0.9 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.3 | 2.4 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.3 | 1.5 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.3 | 1.5 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.3 | 1.7 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.3 | 1.1 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.3 | 0.8 | GO:0001565 | phorbol ester receptor activity(GO:0001565) non-kinase phorbol ester receptor activity(GO:0001566) |
| 0.3 | 0.8 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.3 | 0.8 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.3 | 1.6 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.3 | 1.3 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.3 | 1.3 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.3 | 1.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.3 | 1.0 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.3 | 1.3 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.3 | 1.0 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.3 | 4.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.2 | 0.7 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.2 | 2.5 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.2 | 2.0 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.2 | 2.7 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.2 | 2.4 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.2 | 1.5 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.2 | 2.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 3.7 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.2 | 1.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.2 | 0.9 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.2 | 0.9 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.2 | 1.6 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.2 | 0.9 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.2 | 0.6 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.2 | 0.6 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.2 | 4.9 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.2 | 2.6 | GO:0031386 | protein tag(GO:0031386) |
| 0.2 | 1.0 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.2 | 1.4 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.2 | 1.0 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) |
| 0.2 | 3.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.2 | 0.6 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.2 | 1.4 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.2 | 1.6 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 5.1 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.2 | 0.6 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.2 | 1.5 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.2 | 0.7 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.2 | 3.7 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.2 | 0.7 | GO:0070404 | NADH binding(GO:0070404) |
| 0.2 | 1.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.2 | 1.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.2 | 0.3 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.2 | 2.7 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 1.4 | GO:0046790 | virion binding(GO:0046790) |
| 0.2 | 1.4 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.2 | 3.4 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.2 | 0.5 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.2 | 1.2 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.2 | 0.8 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.2 | 0.6 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.2 | 0.6 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.2 | 2.4 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.2 | 2.6 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.2 | 1.9 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.2 | 1.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.2 | 1.1 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.2 | 1.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 3.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.2 | 0.5 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.2 | 1.4 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.2 | 1.4 | GO:0005536 | glucose binding(GO:0005536) |
| 0.2 | 2.0 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.1 | 1.8 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 3.1 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 1.0 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.1 | 0.9 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.1 | 3.4 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 0.6 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 4.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 1.8 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.6 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.1 | 0.8 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 5.3 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.1 | 9.1 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 0.5 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 2.3 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 0.4 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.1 | 0.5 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.9 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 1.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.4 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 0.7 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 0.8 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.7 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.1 | 0.8 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 1.0 | GO:0032451 | demethylase activity(GO:0032451) |
| 0.1 | 1.0 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.1 | 0.6 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.1 | 3.8 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.1 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.1 | 5.5 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 1.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.9 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 4.5 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.1 | 2.0 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 2.6 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 0.5 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.1 | 3.0 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 2.3 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 1.3 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.1 | 1.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.3 | GO:0046592 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.1 | 0.3 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.1 | 0.4 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.1 | 0.3 | GO:0070538 | oleic acid binding(GO:0070538) lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 2.2 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.1 | 7.3 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.1 | 0.4 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.1 | 1.1 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.1 | 0.6 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 1.0 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 0.9 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.7 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.1 | 1.0 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.1 | 0.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 2.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.3 | GO:0070330 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) aromatase activity(GO:0070330) |
| 0.1 | 1.4 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 6.1 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 1.4 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 1.9 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 1.9 | GO:0008308 | voltage-gated anion channel activity(GO:0008308) |
| 0.1 | 1.8 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.5 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 1.2 | GO:0070636 | purine-specific mismatch base pair DNA N-glycosylase activity(GO:0000701) single-strand selective uracil DNA N-glycosylase activity(GO:0017065) nicotinamide riboside hydrolase activity(GO:0070635) nicotinic acid riboside hydrolase activity(GO:0070636) deoxyribonucleoside 5'-monophosphate N-glycosidase activity(GO:0070694) |
| 0.1 | 0.9 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 0.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.4 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 0.7 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.1 | 0.2 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.1 | 7.5 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 0.7 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 1.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 7.3 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 0.4 | GO:0048038 | quinone binding(GO:0048038) |
| 0.1 | 0.8 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 0.8 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.3 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 0.1 | 1.6 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.1 | 0.8 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.7 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.1 | 0.9 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 1.7 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.9 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 1.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.7 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 1.7 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 1.2 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.1 | 0.9 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 1.1 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 0.6 | GO:0016215 | acyl-CoA desaturase activity(GO:0016215) |
| 0.1 | 4.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 0.2 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.1 | 0.8 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 1.4 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 0.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.4 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.1 | 0.5 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 1.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 1.7 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.1 | 0.2 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.1 | 1.3 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.1 | 1.9 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.4 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.1 | 6.6 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.1 | 1.9 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 1.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 2.4 | GO:0044824 | integrase activity(GO:0008907) T/G mismatch-specific endonuclease activity(GO:0043765) retroviral integrase activity(GO:0044823) retroviral 3' processing activity(GO:0044824) |
| 0.1 | 0.7 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 12.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 0.4 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 0.4 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 1.2 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 0.9 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 0.5 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.3 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.0 | 2.0 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 1.9 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.0 | 0.6 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.2 | GO:0019145 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 1.6 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.2 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.2 | GO:0044020 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 10.4 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.2 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 1.1 | GO:0030546 | receptor activator activity(GO:0030546) |
| 0.0 | 0.6 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 1.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 2.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.8 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 2.1 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 2.7 | GO:0035326 | enhancer binding(GO:0035326) |
| 0.0 | 2.6 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 1.0 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.5 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.5 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 1.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.3 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 0.3 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.0 | 0.4 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 1.4 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.3 | GO:0052890 | acyl-CoA dehydrogenase activity(GO:0003995) oxidoreductase activity, acting on the CH-CH group of donors, with a flavin as acceptor(GO:0052890) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0004549 | tRNA-specific ribonuclease activity(GO:0004549) |
| 0.0 | 0.8 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.3 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0001639 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) |
| 0.0 | 0.4 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.0 | 0.2 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.2 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.9 | GO:0052771 | coenzyme F390-A hydrolase activity(GO:0052770) coenzyme F390-G hydrolase activity(GO:0052771) |
| 0.0 | 0.4 | GO:0035380 | C-3 sterol dehydrogenase (C-4 sterol decarboxylase) activity(GO:0000252) mevaldate reductase activity(GO:0004495) gluconate dehydrogenase activity(GO:0008875) epoxide dehydrogenase activity(GO:0018451) 5-exo-hydroxycamphor dehydrogenase activity(GO:0018452) 2-hydroxytetrahydrofuran dehydrogenase activity(GO:0018453) acetoin dehydrogenase activity(GO:0019152) phenylcoumaran benzylic ether reductase activity(GO:0032442) D-xylose:NADP reductase activity(GO:0032866) L-arabinose:NADP reductase activity(GO:0032867) D-arabinitol dehydrogenase, D-ribulose forming (NADP+) activity(GO:0033709) (R)-(-)-1,2,3,4-tetrahydronaphthol dehydrogenase activity(GO:0034831) 3-hydroxymenthone dehydrogenase activity(GO:0034840) very long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0035380) dihydrotestosterone 17-beta-dehydrogenase activity(GO:0035410) (R)-2-hydroxyisocaproate dehydrogenase activity(GO:0043713) L-arabinose 1-dehydrogenase (NADP+) activity(GO:0044103) L-xylulose reductase (NAD+) activity(GO:0044105) 3-ketoglucose-reductase activity(GO:0048258) D-arabinitol dehydrogenase, D-xylulose forming (NADP+) activity(GO:0052677) |
| 0.0 | 0.2 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.8 | GO:0001076 | transcription factor activity, RNA polymerase II transcription factor binding(GO:0001076) |
| 0.0 | 1.5 | GO:0030792 | methylarsonite methyltransferase activity(GO:0030792) |
| 0.0 | 0.3 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.0 | 0.7 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.4 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.2 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.1 | GO:0070290 | phospholipase D activity(GO:0004630) N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 1.1 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 11.6 | GO:0044822 | poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.3 | GO:0016835 | carbon-oxygen lyase activity(GO:0016835) |
| 0.0 | 0.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0052872 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) phenanthrene 9,10-monooxygenase activity(GO:0018636) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) tocotrienol omega-hydroxylase activity(GO:0052872) thalianol hydroxylase activity(GO:0080014) |
| 0.0 | 0.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.3 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |