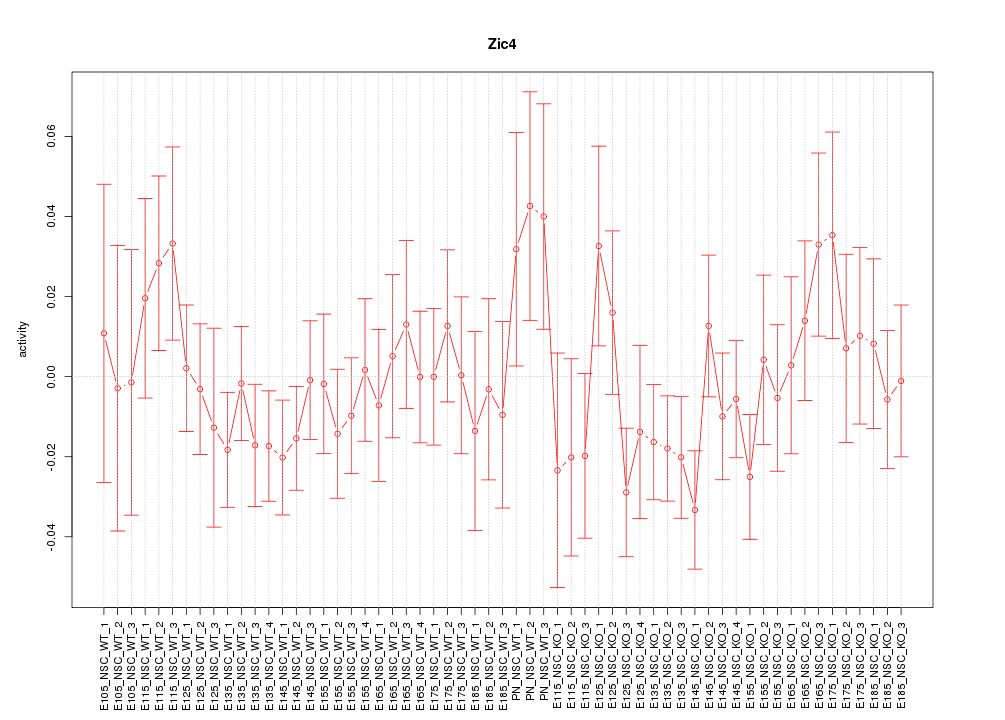
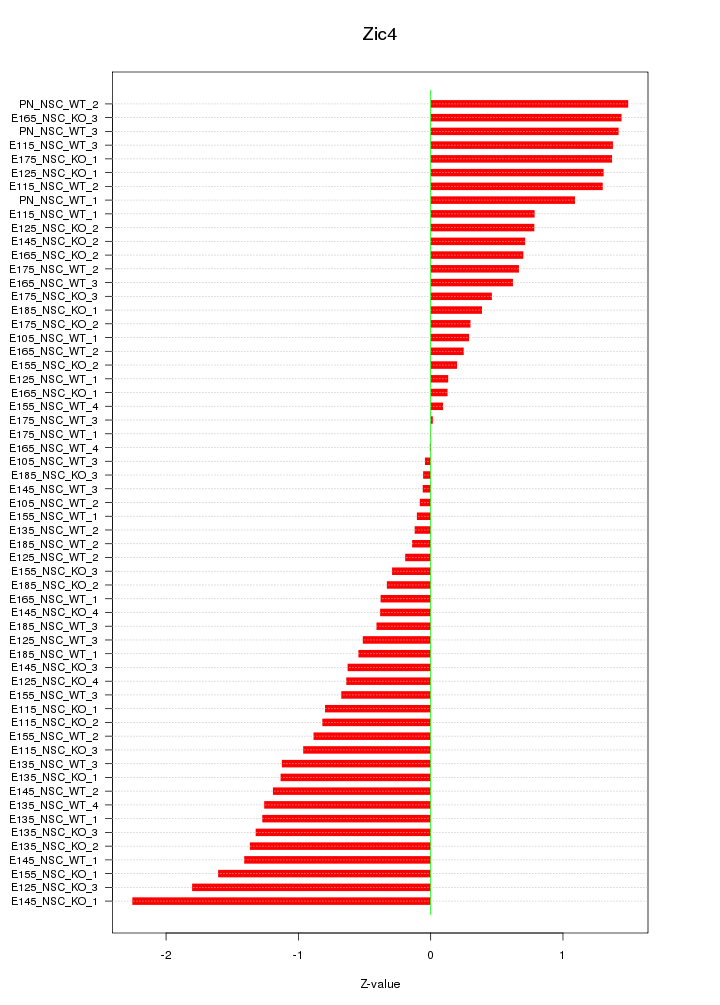
Motif ID: Zic4
Z-value: 0.899
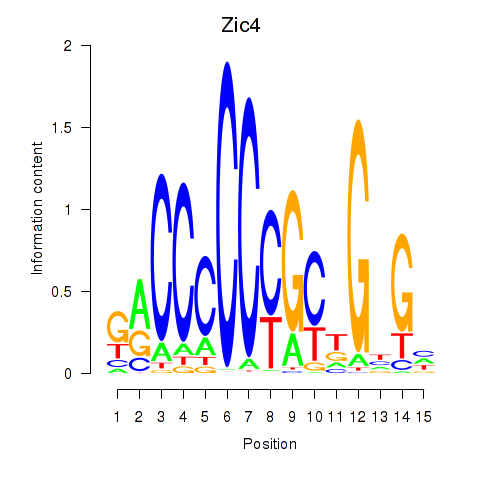
Transcription factors associated with Zic4:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Zic4 | ENSMUSG00000036972.8 | Zic4 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zic4 | mm10_v2_chr9_+_91378636_91378646 | -0.10 | 4.3e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:0061743 | motor learning(GO:0061743) |
| 1.1 | 4.3 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 1.0 | 1.0 | GO:0072554 | blood vessel lumenization(GO:0072554) |
| 0.9 | 4.5 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.8 | 2.3 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.7 | 2.2 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.7 | 3.7 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.7 | 3.7 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.7 | 3.6 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.7 | 2.0 | GO:0060854 | patterning of lymph vessels(GO:0060854) |
| 0.6 | 1.9 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.6 | 1.9 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.6 | 5.8 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.6 | 2.8 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.5 | 2.1 | GO:1903802 | positive regulation of arachidonic acid secretion(GO:0090238) L-glutamate(1-) import into cell(GO:1903802) L-glutamate import into cell(GO:1990123) |
| 0.5 | 3.6 | GO:0070458 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.5 | 2.5 | GO:0030091 | protein repair(GO:0030091) |
| 0.4 | 1.2 | GO:0030070 | insulin processing(GO:0030070) |
| 0.4 | 1.1 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.4 | 1.5 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.3 | 0.9 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) alveolar secondary septum development(GO:0061144) |
| 0.3 | 0.9 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.3 | 1.1 | GO:0061428 | positive regulation of oocyte development(GO:0060282) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.3 | 1.9 | GO:0032811 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) negative regulation of epinephrine secretion(GO:0032811) |
| 0.3 | 4.6 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.3 | 1.0 | GO:0097168 | condensed mesenchymal cell proliferation(GO:0072137) mesenchymal stem cell proliferation(GO:0097168) |
| 0.3 | 0.8 | GO:0097274 | urea homeostasis(GO:0097274) |
| 0.2 | 5.9 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.2 | 1.2 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.2 | 2.7 | GO:0010748 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.2 | 0.7 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.2 | 2.7 | GO:0042711 | maternal behavior(GO:0042711) |
| 0.2 | 0.6 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) regulation of sodium:potassium-exchanging ATPase activity(GO:1903406) |
| 0.2 | 1.4 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.2 | 1.0 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.2 | 1.1 | GO:0002317 | plasma cell differentiation(GO:0002317) negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.2 | 0.9 | GO:1901678 | iron coordination entity transport(GO:1901678) |
| 0.2 | 0.9 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.2 | 1.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.6 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 | 0.6 | GO:0060838 | lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.1 | 0.7 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.8 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 0.3 | GO:0009838 | abscission(GO:0009838) positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.1 | 0.5 | GO:0002182 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.1 | 0.4 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.3 | GO:0015920 | regulation of phosphatidylcholine catabolic process(GO:0010899) lipopolysaccharide transport(GO:0015920) |
| 0.1 | 1.9 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.1 | 1.0 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 1.8 | GO:1903861 | regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 0.2 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.1 | 0.1 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.1 | 0.5 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.1 | 1.6 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.1 | 0.4 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.1 | 0.5 | GO:0031424 | keratinization(GO:0031424) establishment of skin barrier(GO:0061436) |
| 0.1 | 7.8 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 0.7 | GO:0032438 | melanosome organization(GO:0032438) endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.0 | 0.6 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 1.6 | GO:0060351 | cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.0 | 0.3 | GO:0006549 | isoleucine metabolic process(GO:0006549) branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 | 0.3 | GO:0003161 | cardiac conduction system development(GO:0003161) |
| 0.0 | 0.1 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.0 | 0.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.2 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.2 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.9 | GO:0060441 | epithelial tube branching involved in lung morphogenesis(GO:0060441) |
| 0.0 | 0.7 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.1 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.0 | 0.2 | GO:0070234 | positive regulation of T cell apoptotic process(GO:0070234) |
| 0.0 | 0.9 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 0.1 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.1 | GO:1900736 | regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900736) |
| 0.0 | 0.3 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.0 | 0.8 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.8 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 1.9 | GO:0007369 | gastrulation(GO:0007369) |
| 0.0 | 0.2 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.4 | GO:0002092 | positive regulation of receptor internalization(GO:0002092) |
| 0.0 | 2.9 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.0 | 1.5 | GO:0045598 | regulation of fat cell differentiation(GO:0045598) |
| 0.0 | 0.4 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.8 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.7 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.7 | 2.7 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.6 | 3.7 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.5 | 2.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.4 | 1.9 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.3 | 4.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 1.1 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.2 | 2.7 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.6 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 0.9 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 3.2 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.4 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 2.4 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 1.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.7 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 2.0 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.3 | GO:0090543 | Flemming body(GO:0090543) |
| 0.1 | 0.8 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 1.0 | GO:0098984 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.1 | 0.6 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 4.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 3.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 2.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.7 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.5 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.6 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 1.1 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 8.0 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.0 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.1 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.3 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 5.2 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.2 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.7 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.8 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 1.1 | 4.5 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.9 | 5.5 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.5 | 1.5 | GO:0005119 | smoothened binding(GO:0005119) hedgehog receptor activity(GO:0008158) hedgehog family protein binding(GO:0097108) |
| 0.4 | 5.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.4 | 4.8 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.3 | 1.9 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.3 | 0.9 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 0.3 | 1.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.3 | 3.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.3 | 1.1 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) tubulin deacetylase activity(GO:0042903) |
| 0.3 | 2.0 | GO:0001224 | RNA polymerase II transcription cofactor binding(GO:0001224) |
| 0.3 | 2.4 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.2 | 2.5 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.2 | 0.7 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) oncostatin-M receptor activity(GO:0004924) |
| 0.2 | 0.8 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.2 | 0.6 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.2 | 2.2 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.2 | 2.3 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.2 | 4.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 1.0 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.2 | 1.9 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.1 | 9.8 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 3.6 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.1 | 2.1 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 0.9 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.1 | 2.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 0.4 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.3 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 0.2 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.1 | 0.6 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 1.1 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.1 | 0.7 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 0.3 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.5 | GO:0043023 | GTPase inhibitor activity(GO:0005095) ribosomal large subunit binding(GO:0043023) |
| 0.0 | 3.4 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 2.8 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.0 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 3.1 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.3 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 1.9 | GO:0004120 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) photoreceptor cyclic-nucleotide phosphodiesterase activity(GO:0004120) 7,8-dihydro-D-neopterin 2',3'-cyclic phosphate phosphodiesterase activity(GO:0044688) inositol phosphosphingolipid phospholipase activity(GO:0052712) inositol phosphorylceramide phospholipase activity(GO:0052713) mannosyl-inositol phosphorylceramide phospholipase activity(GO:0052714) mannosyl-diinositol phosphorylceramide phospholipase activity(GO:0052715) |
| 0.0 | 0.7 | GO:0016917 | GABA receptor activity(GO:0016917) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.4 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 1.0 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.8 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 1.1 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 2.4 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 1.1 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 1.0 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 1.2 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 1.7 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.3 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.0 | 0.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.2 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.4 | GO:0008748 | N-ethylmaleimide reductase activity(GO:0008748) reduced coenzyme F420 dehydrogenase activity(GO:0043738) sulfur oxygenase reductase activity(GO:0043826) malolactic enzyme activity(GO:0043883) epoxyqueuosine reductase activity(GO:0052693) |